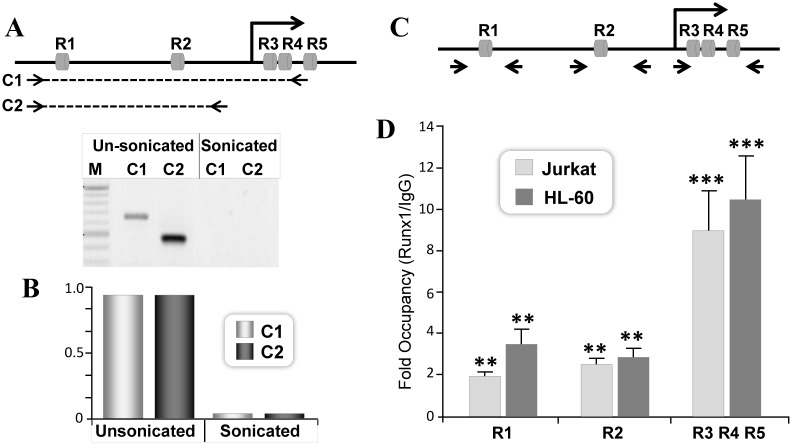
Fig 4. Endogenous RUNX1 protein is recruited to RUNX motifs located in the P1 promoter and the 5’UTR.
A) Efficiency of chromatin shearing of the RUNX1 locus in Jurkat cells. Control (unsonicated) and sheared (sonicated) chromatin was subjected to PCR using either (A) traditional or (B) real time qPCR analysis. Primer pair amplifying RUNX1 chromatin loci encompassing R1-R4 (C1) or R1-R2 (C2) is indicated with dashed lines. The amplified 650bp C1 and 500bp C2 region of the RUNX1 gene were detected in control but not in sonicated chromatin. M denote 100bp ladder. B) Lack of intact DNA between the promoter and the UTR RUNX sites in sonicated chromatin was confirmed by real time qPCR. C) Arrowhead indicates relative position of primer pairs used to assess RUNX1 occupancy on promoter and UTR RUNX motifs D) Equivalent amount of chromatin (3U OD260) from Jurkat or HL-60 cells were used for ChIP assays. Chromatin was immunoprecipitated with 2μg of IgG or RUNX1 polyclonal antibody. Site specific occupancy was determined by real time PCR. Mean values of IgG or RUNX1 immunoprecipitated DNA from three independent experiments with three replicates were obtained for each site. Data is expressed as fold changes over IgG in respective cells. Asterisks indicate statistically significant enrichment of DNA immunoprecipitated with RUNX1 antibody (**p<0.01, ***p<0.001).