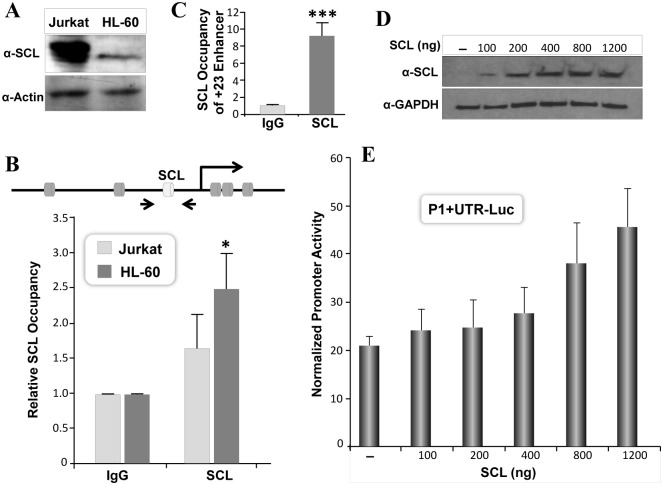
Fig 6. SCL transcription factor activates RUNX1-P1 promoter.
A) Equal amount of total protein from Jurkat and HL-60 cells were resolved by SDS-PAGE. Blots were probed with polyclonal SCL antibody, stripped and reprobed for tubulin antigen, used as a loading control. B) Jurkat and HL-60 cells were processed for chromatin immunoprecipitation. SCL occupancy of SCL motif in the RUNX1 P1 promoter was determined by qPCR. The positions of the SCL motif and the primer pair for ChIP assays are indicated in the diagram. Average data from three independent ChIP experiments, with three replicate each is shown as relative to IgG. C) Jurkat cells were processed for ChIP assays exactly as described in (B). Occupancy of SCL motif present in +23 RUNX1 enhancer was determined by qPCR. Average data from three independent ChIP experiments, with three replicate each is shown as relative to IgG. Statistical significance of SCL occupancy is indicated by asterisks (***p<0.001). D) HeLa cells were transiently co-transfected with RUNX1 promoter and indicated amounts of SCL expression vector. Cells were harvested 18h later for western blot analysis. SCL protein was detected by probing blots with SCL antibody. GAPDH is shown as internal loading control in the same blots. E) HeLa cells were co-transfected with 200ng of the RUNX1 P1+UTR Luc and increasing concentration of SCL expression vector. Luciferase activity was determined 18h later and normalized with Renilla luciferase. Bars in graph represent mean values of three independent experiments each performed in triplicate.