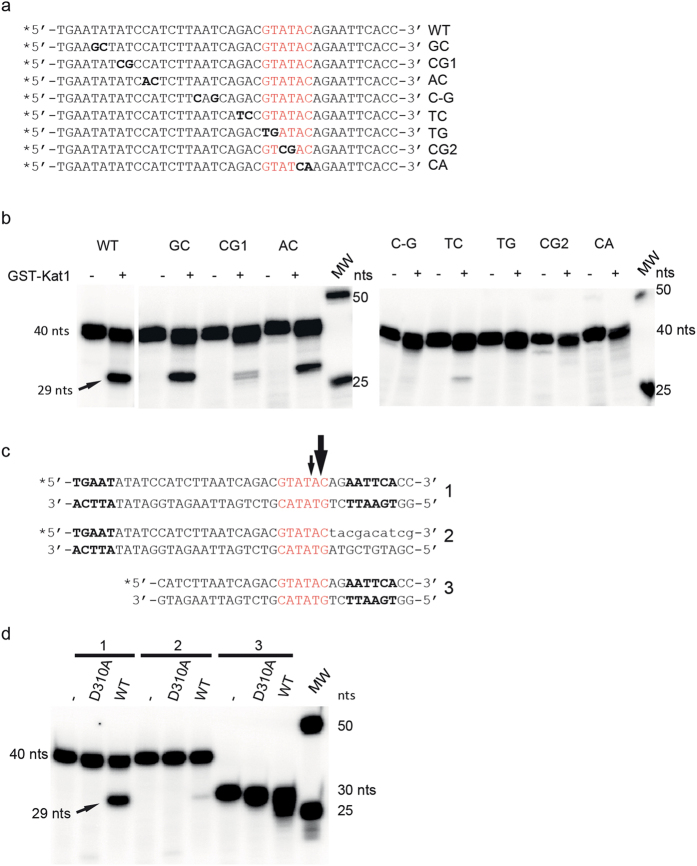
Figure 2. The Kat1 recognition site in TIR-R extends beyond the GTATAC sequence.
(a) DNA sequences showing the upper labeled strand of 40-bp duplexes containing mutations in TIR-R. Mutations are indicated in bold letters and designated WT, GC etc. The GTATAC sequence at the beginning of TIR-R is indicated in red. (b) The indicated mutant DNA substrates were incubated with Kat1 (+) or without protein (−) using the conditions described in Fig. 1, except that the gel was thicker. The 40-nt substrates, the 29-nt products and DNA ladder (MW) is shown. (c,d) The Kat1 endonuclease activity of TIR-R requires a flanking DNA sequence. (c) Representation of DNA duplexes used as substrates, the WT 40-bp duplex (1), a 40-bp duplex with 10-bp scrambled on the flanking DNA end (2) and a 30-bp duplex (3). Arrows indicate incisions on the top strand, asterisks positions of 32P-label (upper strand was labeled) and bold letters indicate subterminal repeats. (d) Nuclease assay using WT Kat1, Kat1D310A and no protein (−). Reaction conditions were as described in Fig. 1. The radiogram depicts the 40-nt and 30-nt substrates, the 29-nt product and size markers. Note that both the 30-bp substrate and 40-bp substrate with a scrambled host end lack subterminal repeats TGAAT and AATTCA, respectively.