Fig. 5.
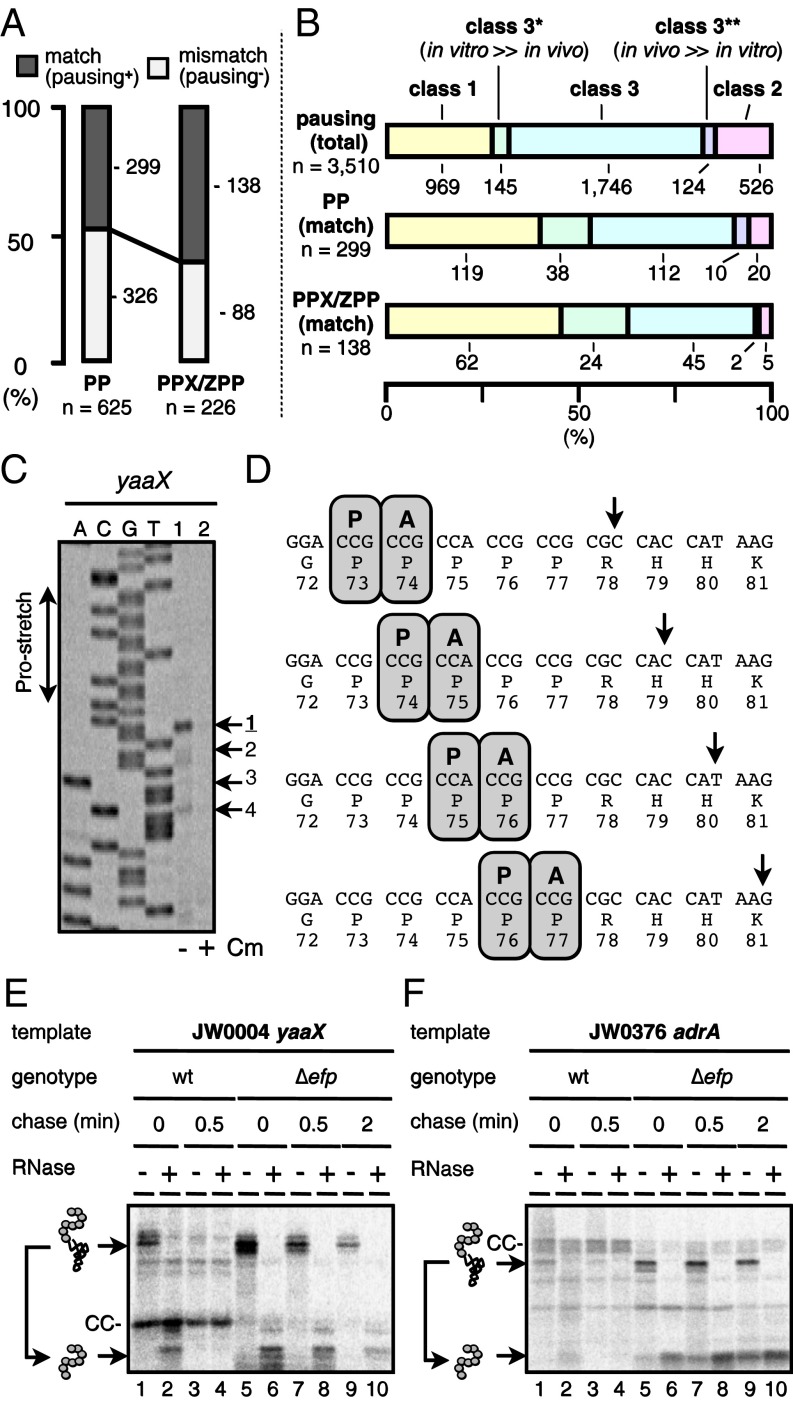
Detection of proline-based pausing by iNP. (A) The occurrence of pausing around the 625 PP motifs and the 226 PPX/ZPP motifs in the 1,038 target genes. The existing PP and PPX/ZPP motifs were scored by whether they were (match) or were not (mismatch) accompanied by the accumulation of polypeptidyl–RNAs, whose estimated sizes can be accounted for by pausing therein (Materials and Methods). (B) Classification of the putative PP- and PPX/ZPP-mediated pausing events. The classification of the 299 PP-matched pausing events and the 138 PPX/ZPP-matched pausing events that were assigned as proline-mediated in A into the class 1, class 2, or class 3 (denoted “both”) categories is compared with the equivalent classification of the 3,510 total pausing events. In addition, distributions into subclasses of class 3 are included: class 3*, normalized polypeptidyl–tRNA intensity higher in vitro than in vivo by a factor of 3 or more; class 3**, normalized polypeptidyl–tRNA intensity higher in vivo than in vitro by a factor of 3 or more. (C) Toeprint confirmation of ribosome stalling on yaaX. PURE system translation complexes directed by the yaaX template in the absence (lane 1) or presence (lane 2) of chloramphenicol were subjected to the toeprint assay. Dideoxy sequencing products of yaaX are shown in lanes A, C, G, and T. (D) Schematic illustration of ribosome pausing on the yaaX mRNA. (E and F) Effects of the ∆efp mutation on translation pausing observed with yaaX and adrA. ASKA library clones of yaaX (E) and adrA (F) were expressed in the original host (efp+: lanes 1–4) and in its ∆efp derivative (lanes 5–10). Cells were pulse-labeled with [35S]methionine for 0.5 min and then were chased with unlabeled methionine for the indicated periods (0, 0.5, or 2 min). Samples were processed by the in vivo iNP procedures.