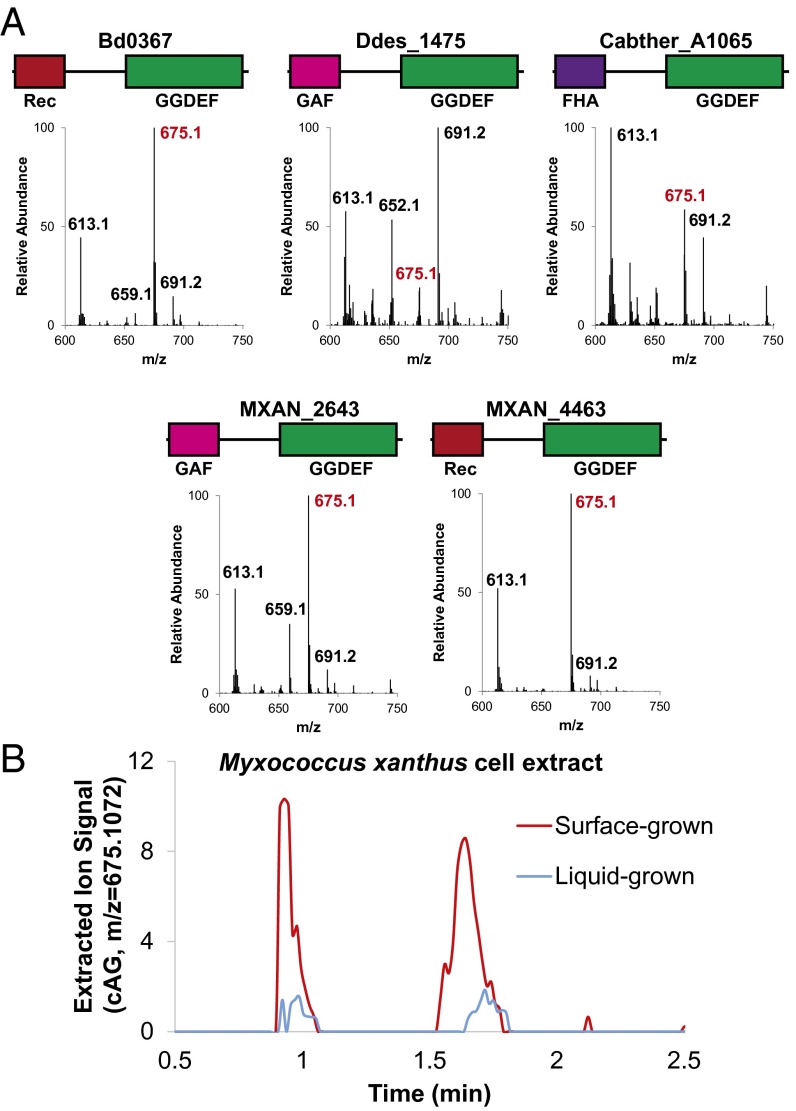
Fig. 4.
Validation of Hypr activity in select Deltaproteobacteria and Acidobacteria. (A) LC/MS analysis of E. coli cell extracts overexpressing candidate Hypr enzymes; see SI Appendix, Fig. S1 for protein domain color scheme and SI Appendix, Fig. S12 for corresponding protein gel. Bd, Bdellovibrio bacteriovorus; Ddes, Desulfovibrio desulfuricans; Mxan, Myxococcus xanthus; Cabther, Candidatus Chloracidobacterium thermophilum. Shown are the MS spectra from integrating the retention time region containing all three cyclic dinucleotides. Expected masses are for cdiG (m/z = 691), cAG (m/z = 675), and cdiA (m/z = 659). (B) LC/MS analysis of M. xanthus cell extracts from surface- or liquid-grown samples. Shown is the extracted ion trace for cAG (m/z = 675.1072; ppm < 10 cutoff) normalized to the weight of extracted cells. A second biological replicate is shown in SI Appendix, Fig. S15.