Fig 1. Liver development in the maternal-zygotic leg1azju1 mutant is amenable to the environmental changes.
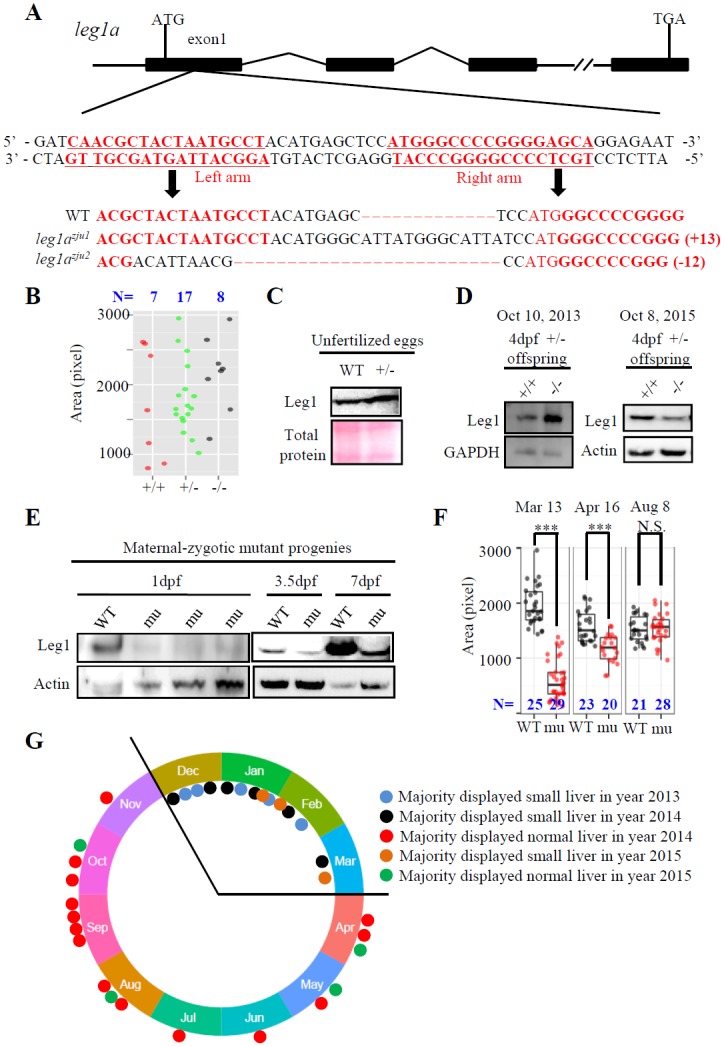
(A) Top panel: Schematic diagram showing the genomic structure of the leg1a gene. Black box: exons; solid line: introns; double slashes: omitted genomic region. Middle panel: The left and right TALEN targeting sequences in the exon1 are lettered in red. Bottom panel: Comparison of the genomic DNA sequence among WT, leg1azju1 (with 13 bp insertion) and leg1azju2 (with 12 bp deletion) two mutant alleles. TALEN target sequences are in red. (B-D) The leg1azju1 homozygotes (-/-) obtained from a cross between heterozygous (+/-) parents showed normal liver development (B). Each dot represents the liver size (measured based on the signal area of fabp10a) of a single embryo. Three independent experiments were carried and a representative one is shown here. Total Leg1 was detected in unfertilized eggs (C) and in two independent samples of 4-dpf WT (+/+) and mutant (-/-) embryos collected at different dates (D). (E-G) The maternal-zygotic leg1azju1 homozygotes (mu) obtained from a cross between homozygous parents lacked Leg1 at 1 dpf, but expressed Leg1b at 3.5 dpf and 7 dpf (E), and showed a small liver phenotype on March 13, an intermediate-sized liver on April 16, and a normal sized liver on August 8, 2015. (F). Recordings of liver phenotype in 32 cases from December 30 2013 to October 4 2015 showed majority of the maternal-zygotic leg1azju1 homozygotes (mu) exhibited a small liver in 14 cases recorded in cold seasons but a big variation in 18 cases recorded in warm/hot seasons (G). ***, p<0.001, N.S., not significant. Western blot was repeated three times for (C), five times for (D) and (E).
