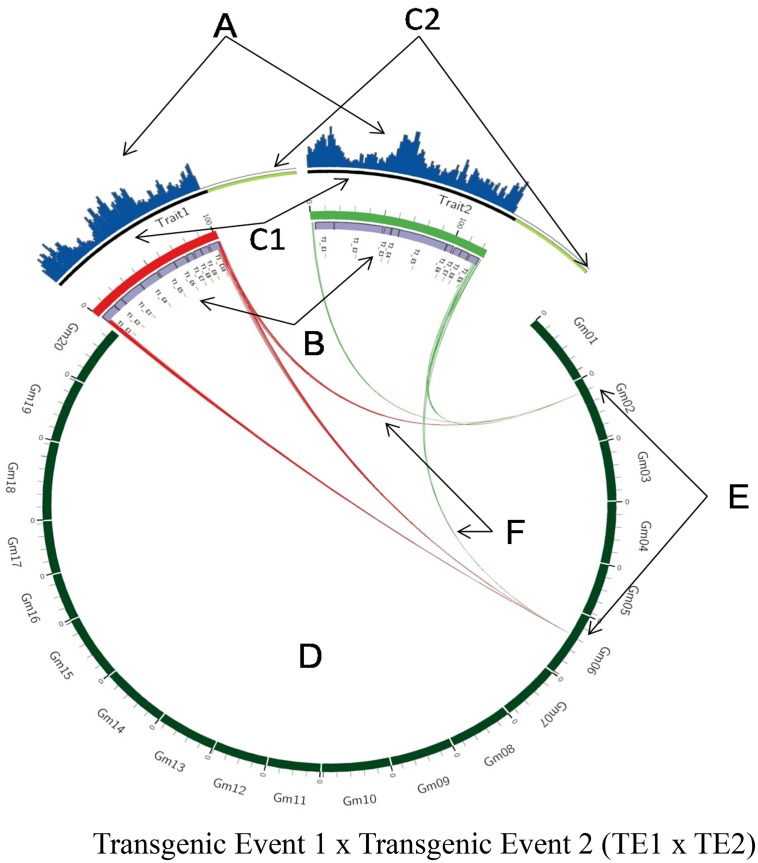
Fig 3. Molecular characterization of soybean breeding stack Transgenic Event 1 x Transgenic Event 2 (TE1 x TE2) using whole genome sequencing.
Genomic DNA of TE1 x TE2 was randomly sheared and sequenced using Illumina's HiSeq2000 instrument. The genome coverage was ~ 14X, i.e. 14 copies of soybean haploid genome. Short HiSeq2000 reads (A) spanning entire T-DNA within TE1 and TE2 (B) were mapped back to transformation plasmid that contained intended T-DNA (C1) and backbone (C2). Uninterrupted blue bars aligned to the intended T-DNA (C1) of the transformation plasmid confirms the integrity of T-DNA within TE1 and TE2. No blue bars over plasmid backbone (C2) confirms the absence of those sequences within the genome of TE1 and TE2. Twenty chromosomes (Gm1-20) of soybean reference genome (Williams 82 version X) are represented in circular fashion (D). Reads spanning junction regions were mapped back to soybean reference genome, which showed single insertion site on chromosome 6 (E) in TE1 and on chromosome 2 (E) in TE2. T-DNA insert in both TE1 and TE2 share the same fragment at the 3'border region (F).