Fig. 1. Phylogeny and photoactivity of G. theta ACRs.
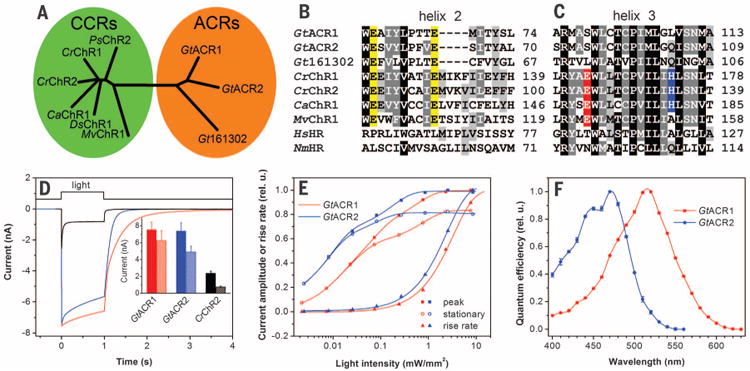
(A) Phylogenetic tree of CCRs and ACRs. (B and C) ClustalW alignments of transmembrane helices 2 (B) and 3 (C) Abbreviated organism names are: Gt Guillardia theta; Cr Chlamydomonas reinhardtii; Ca Chlamydomonas augustae; Mv Mesostigma viride; Hs, Halobacterium salinarum; Nm, Nonlabens marinus. The last residue numbers are shown on the right. Conserved Glu residues in helix 2 are highlighted in yellow, Glu residues in the position of bacteriorhodopsin Asp85 in red, and His residues corresponding to His134 of CrChR2 in blue. (D) Photocurrents of GtACR1, GtACR2, and CrChR2 in HEK293 cells in response to a saturating light pulse at −60 mV. (Inset) Mean amplitudes of peak (solid bars) and stationary (hatched bars) currents (n = 18 to 20 cells). (E) Dependence of the peak and stationary current amplitudes and rise rates on stimulus intensity. (F) Action spectra of photocurrents.
