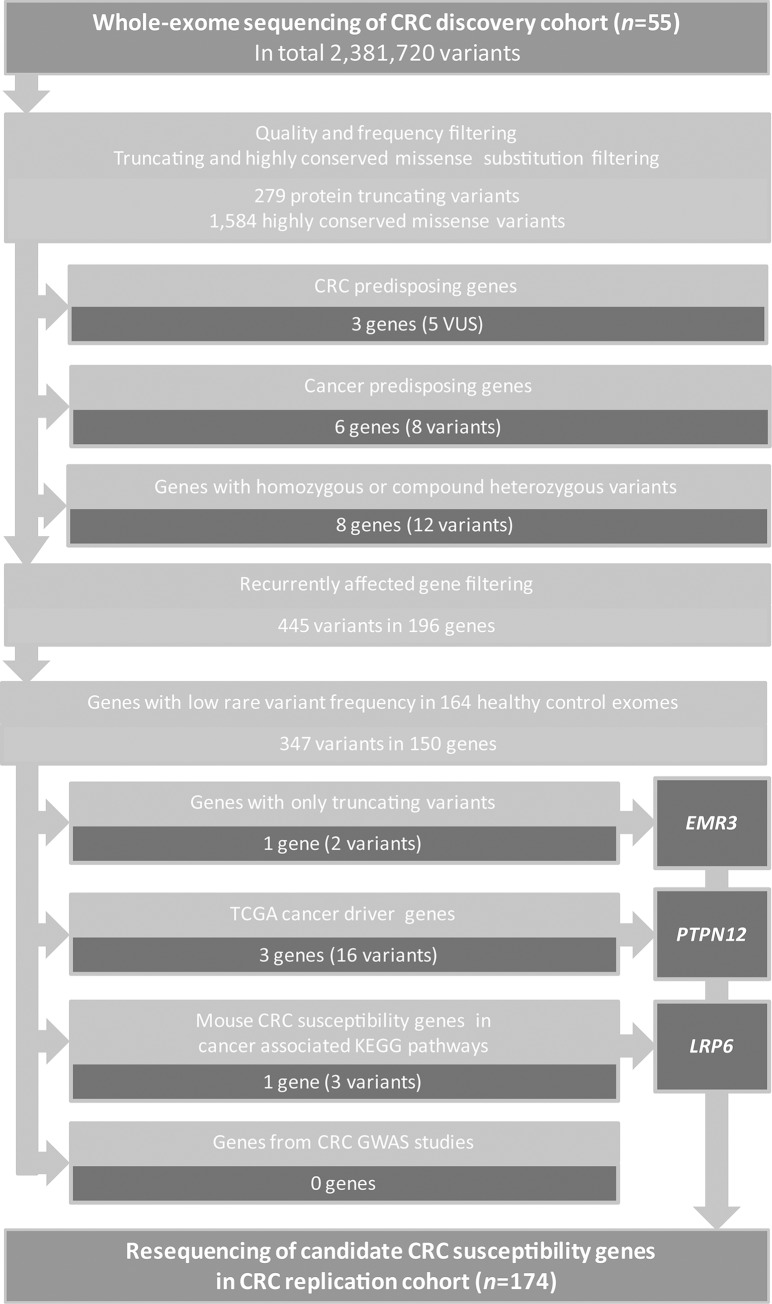
Fig 1. Study design, variant filtering and candidate gene prioritization.
Whole-exome sequencing was performed on germline DNA of 55 early-onset CRC cases. The exome data were first filtered for quality and frequency, followed by filtering for protein truncating and highly conserved missense variants. Next, we removed all known loss-of-function-tolerant genes from this list and searched for known and novel CRC predisposing gene variants.[12,13,57] An additional filtering was applied to identify genes that were affected by two or more potentially pathogenic variants and to remove genes that are frequently affected by protein-truncating or highly conserved missense variants in healthy controls. The remaining set of recurrent variants was filtered for (i) genes recurrently affected by protein truncating variants; (ii) cancer driver genes in CRC [23]; (iii) genes identified as CRC susceptibility genes in mice [29,30] and involved in cancer-related KEGG pathways [hsa04310 (WNT signaling), hsa04350 (TGF-beta signaling), hsa03430 (mismatch repair), hsa03410 (base excision repair), hsa03420 (nucleotide excision repair), map03450 (non-homologous end-joining), hsa03460 (Fanconi anemia), and hsa05200 (pathways in cancer)] (iv) genes identified in CRC GWAS studies [5–7,32]. Genes that remained after these filter steps were selected for re-sequencing in a replication cohort of 174 CRC cases. CRC: colorectal cancer; VUS: variant of unknown significance.