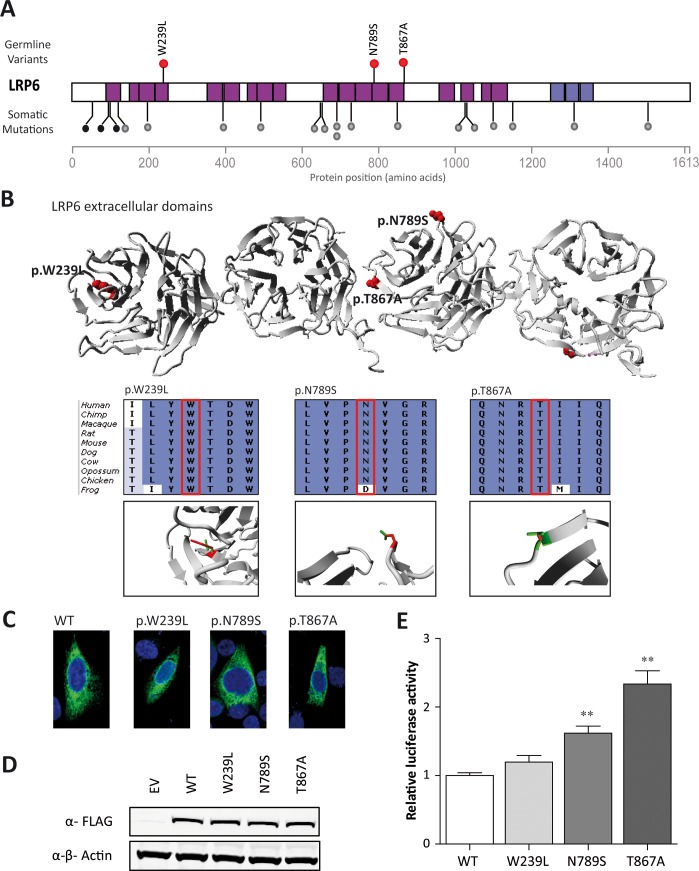
Fig 3. Rare variants in LRP6 in three cases.
(A) Distribution of missense LRP6 variants identified in the CRC discovery cohort (red dots). Somatic LRP6 mutations identified in colorectal tumors [24] are indicated with grey (missense) and black (protein truncating) dots. Structural domains include the β-propeller domains that are used to form the receptor complex (pink bars), and the transmembrane domain (purple). (B) 3D protein structure of the β-propeller domains of LRP6 with the positions of the identified missense variants in red. Insets show conservation of the region in which the missense variants (indicated with the red box) are located with, underneath, close ups of the local 3D protein structure with mutant (red) and wild-type (green) residues. The mutant residue at position 239 is predicted to disturb the protein structure (project HOPE; http://www.cmbi.ru.nl/hope/). The mutant residue at position 789 is much smaller than the wild-type residue and may disturb the binding of Dickkopf-1. Residue 867 is located on the surface of the protein and the mutant residues are not expected to disturb protein structure, but may influence protein binding. (C) Immunofluorescence analyses of LRP6 wild-type and mutant proteins showing similar subcellular localizations. (D) LRP6 protein expression levels normalized to β-actin are similar between wild-type and mutant LRP6. (E) TOPflash analyses of wild-type and mutant LRP6 to determine their effects on the WNT signaling pathway. Luciferase activity was normalized to control and wild-type constructs. Both p.N789S and p.T867A mutants reveal a significant increase in activation compared to the wild-type LRP6 protein. Experiments were performed three times in triplicate. **P <0.001; error bars represent the standard error of the mean.