FIGURE 3.
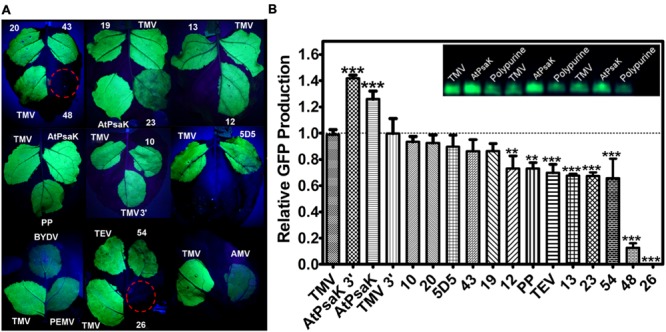
Evaluation of diverse 5′ UTRs on GFP production. Leaves of N. benthamiana were agroinfiltrated with BeYDV containing different 5′ UTRs upstream from the GFP gene vectors (vector pBYR2eXX-GFP, where XX denotes the individual 5′ UTRs). (A) Leaves were photographed at 4 DPI under UV illumination (365 nm). Images are representative of 3–4 independently infiltrated leaves. (B) Agroinfiltrated leaves were harvested between 4 and 5 DPI and extracts were analyzed by SDS-PAGE followed by observation under UV illumination (365 nm) and Coomassie staining. GFP band intensity was quantified using ImageJ software, using native plant proteins as a loading control. Columns represent means ± standard error of three or more independently infiltrated leaves. All leaves were infiltrated with the TMV 5′ UTR vector in addition to the other vectors as an internal control for leaf and plant variability. Two stars (∗∗) indicate p < 0.05 and three stars (∗∗∗) indicate p < 0.01 as compared to TMV by Student’s t-test. 5′ UTR key (position -1 taken as first nucleotide upstream from ATG): TMV, tobacco mosaic virus full length 5’ UTR; AtPsaK 3′, nucleotides -1 to -41 of AtPsaK gene; AtPsaK, nucleotides -1 to -63 of AtPsaK gene; TMV 3′, nucleotides -1 to -21 of TMV; TEV, tobacco etch virus full length 5′ UTR; PP, synthetic polypurine sequence; AMV, full length alfalfa mosaic virus 5′ UTR; BYDV, full length barley yellow dwarf virus 5′ and 3′ UTRs; PEMV, full length pea enation mosaic virus RNA 2 5′ and 3′ UTRs; 10; 20; 5D5; 43; 19; 12; 13; 23; 54; 48; 26, human-derived 5′ UTR sequences.
