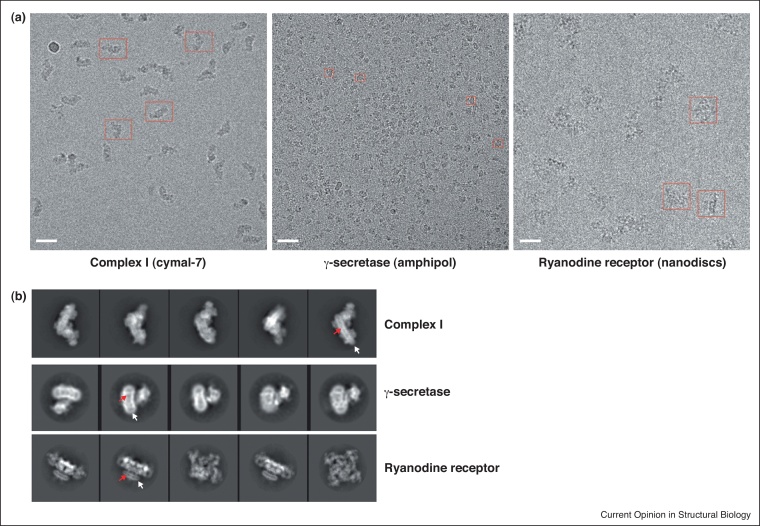
Figure 1.
Membrane protein imaging as single particles by electron cryomicroscopy. (a) Selected areas of micrographs of membrane proteins observed in detergent (Complex I), amphipol (γ-secretase) and nanodisc (ryanodine receptor). The images of Complex I and RyR were taken with the FEI Falcon II detector and the γ-secretase was imaged with the Gatan K2 Summit detector. Scale bar is 400 nm. In the case of RyR receptor, fluorinated octyl maltoside was added to the protein solution just before freezing to get better distribution of the receptor [38•]. The high contrast seen in these images is due to the use of relatively high dose and sufficient defocus. For example, the Complex I images were captured for 4 s (72 frames) at a total dose of ∼68 electrons/Å2. This high dose image was only used for particle picking and contrast transfer function estimation. For subsequent refinements the last 40 frames were discarded [58•]. (b) Reference-free 2D class averages of Complex I, γ-secretase and ryanodine receptor. The panel shows a selection of 2D class averages of different membrane proteins revealing the prominent detergent/lipid belt around the protein (visible as a band around the membrane part and marked with white arrow), transmembrane (TM) helices in the membrane domain (marked with red arrow) and the quaternary structure. In the case of Complex I, the location of some of the supernumerary subunits can be seen in the class averages. The box sizes in the 2D class averages are 280, 90 and 384 pixels for Complex I (1.72 Å/pixel), γ-secretase (2.8 Å/pixel) and ryanodine receptor (1.45 Å/pixel) respectively.