Fig. 1.
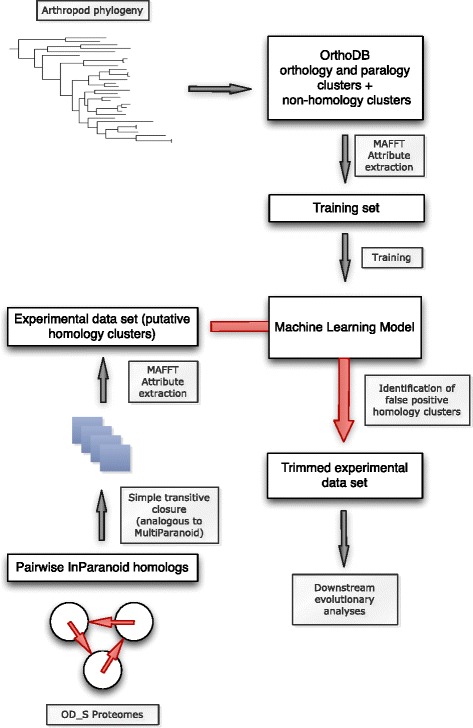
A diagram of the workflow. This figure shows the different steps that were used in developing our machine learning model. Arthropod phylogeny was generated in previous studies and deposited in OrthoDB. These sequences were then gathered from OrthoDB and used as our orthology and paralogy clusters. They were combined with generated non-homology clusters. The combination represents our training data set used to train the machine learning algorithms. The experimental data were assembled with proteins inferred from the assemblies. InParanoid was then used to identify putative homologs. Once putative homologs were identified they were input into the trained machine learning algorithms for classification and subsequent cluster trimming
