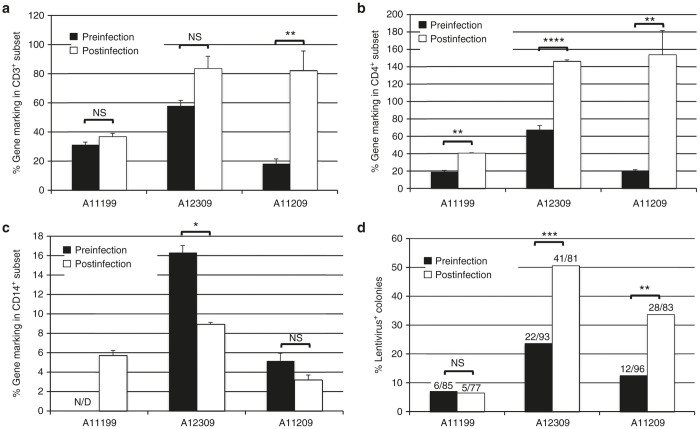
Figure 5.
Gene marking in hematopoietic subsets following Cal-1 transplant and SHIV challenge. Before (200–300 days posttransplant; black bars) or after SHIV challenge (100 days postinfection, white bars), whole blood and bone marrow were collected from the indicated Cal-1-transplanted animal. Bead-based positive selection was used to isolate peripheral blood CD3+ (a), CD4+ (b), and CD14+ cells (c), followed by lentiviral gene marking measurement by Taqman. 100% gene marking represents an average of 1 copy of vector provirus per cell, based on a standard curve constructed from a single-copy lentivirus-infected cell line. Error bars represent standard error of the mean for 2–5 independent Taqman analyses of the same gDNA sample. (d) Bone marrow was hemolysed, and total leukocytes were plated for colony-forming assays. Shown is the percentage of PCR-screened colonies that were positive for actin and lentiviral backbone versus actin alone. Values over each bar represent the number of lentivirus-positive colonies (numerator), as a function of total actin-positive colonies (denominator). P values: NS: not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. SHIV, simian/human immunodeficiency virus.