Abstract
Chemical genomics has the unique potential to expose novel mechanisms of complex cellular biology through screening of small molecules in in vitro assays of a biological phenotype of interest, followed by target identification. In the case of disease-specific assays, the cellular proteins identified might constitute novel drug targets, and the small molecules themselves might be developed as drug leads. In cardiovascular biology, a chemical genomics approach to study the formation of cardiomyocyte, vascular endothelial, and smooth muscle lineages might contribute to therapeutic regeneration. Here, we describe methods used to develop high content screening assays implementing multipotent cardiovascular progenitors derived from human pluripotent stem cells and have identified novel compounds that direct cardiac differentiation.
Keywords: Human pluripotent stem cells, Small molecules, Small RNAs, High content screening, Cardiac differentiation
1 Introduction
The ability to generate pluripotent stem cells (PSC) from humans has provided the unprecedented opportunity to study human development and disease in vitro [1, 2]. Efforts over the past decade to define the mechanisms underlying cardiomyocyte generation from pluripotent stem cells have led to the commercial production of human cardiomyocytes from pluripotent stem cells and have shed light on fundamental developmental mechanisms that may underlie the etiology of congenital heart disease [3, 4]. Moreover, these endeavors might lead to strategies for therapeutic regeneration of adult hearts, which retain a modest ability to self-renew following injury [5].
To probe the biology of PSC differentiation to the cardiac lineage in a large-scale and unbiased way, we have developed phenotypic high throughput assays that allow the simultaneous screening of thousands of bioactive compounds. The large amount of data points in such screens accelerates the discovery of novel biological mechanisms relevant for stem cell differentiation. A typical screening assay consists of large-scale expansion of PSC, which are differentiated in bulk to enrich for a progenitor of interest. Progenitor enriched cultures are then seeded into a high throughput plate format, after which small molecules can be added at any time. The effects of compounds are visualized through a reporter system, usually based on phenotype specific promoters driving a fluorescent or luciferase reporter, or through staining with fate specific antibodies. High throughput plate readers or microscope systems are then implemented to collect data. Consecutive steps comprise hit verification, analysis of the biological mode of action and target identification of the discovered small molecules.
Our screening campaigns using differentiating PSC cultures have led to the discovery of two important classes of molecules that drive the conversion of mesoderm to cardiac mesoderm via TGFβ inhibition and cardiac mesoderm to cardiomyocytes through Wnt inhibition [6–9]. Stem cell-based screening approaches are however not limited to cardiac differentiation, for example small molecules targeting endoderm differentiation have been identified using similar screening methods [10, 11]. Here, we provide methods for chemical genomics applied to cardiovascular biology, which has already yielded insights into basic differentiation mechanisms and reagents to generate large numbers of pure cardiomyocytes. Such molecules might moreover be useful as tools to probe whether the target proteins can be engaged to enhance the limited regenerative potential of the adult heart [1, 4, 12].
2 Materials and Equipment
2.1 Reagents
Human PSC (hPSC) Growth medium: Knockout Dulbecco's Modified Eagle's medium (KO DMEM, Invitrogen) supplemented with 2 mM l-glutamine with 20 % Knockout Serum Replacement (KOSR, Invitrogen), 0.1 mM nonessential amino acids (NEAA), 50 U/mL penicillin, 50 mg/mL streptomycin, 0.1 mM beta-mercaptoethanol, and 8 ng/mL basic fibroblast growth factor (bFGF). An alternative medium can be used to maintain hPSC in culture (see Note 1).
hPSC Differentiation medium: Stem Pro 34 with the included supplement and 2 mM l-glutamine, 0.1 mM NEAA, 50 U/mL penicillin, 50 mg/mL streptomycin, 0.1 mM beta-mercaptoethanol, and 5 ng/mL ascorbic acid.
Mouse embryonic fibroblast (MEF) medium: DMEM High Glucose (4.5 mg/mL), 10 % Fetal Bovine Serum (FBS), 2 mM l-glutamine, 50 U/mL penicillin, 50 mg/mL streptomycin.
Serum-free medium (SFM): 75 % Iscove's Modified Dulbecco's Medium (IMDM), 25 % Ham's-F12 medium complemented with 2 mM l-glutamine, 50 U/mL penicillin, 50 mg/mL streptomycin, 1 % of the serum replacing B27 supplement without vitamin A (GIBCO), 0.5 % of the serum replacing N2 supplement (GIBCO), 0.5 mM ascorbic acid, 0.05 % Bovine Serum Albumin (BSA), and 0.46 mM 1-thioglycerol. This medium is stable for about a week and is ideally made fresh for every use.
1× TrypLE Express dissociation reagent (GIBCO).
Collagenase IV: diluted in KO DMEM at 1 mg/mL or 1.5 mg/mL (concentration depends on the application).
83.3 μg/mL Growth factor reduced Matrigel (BD Biosciences): diluted in ice cold KO DMEM.
0.1 % Gelatin solution: diluted in sterile water.
10 ng/μL Activin A (R&D Systems): diluted in Phosphate Buffered Saline (PBS) with 0.1 % BSA (see Note 2).
25 ng/μL Bmp4 (R&D Systems): diluted in PBS containing 0.1 % BSA and 4 mM HCl (see Note 2).
25 ng/μL bFGF: diluted in KO DMEM.
10 mM Inhibitor of Wnt Response (IWR): 53AH [6] or the analog IWR-1 diluted in dimethylsulfoxide (DMSO).
30 μM Triiodothyronine (T3): diluted in DMEM high glucose (see Note 3).
Opti-MEM (GIBCO) (optional, if screening siRNA instead of small molecules) (see Note 4).
Lipofectamine RNAiMax (Invitrogen) (optional, if screening siRNA or miRNA instead of small molecules) (see Note 4).
10 mM Thiazovivin: diluted in DMSO.
Ultralow attachment 6-well plates (CoStar).
0.1 % gelatin/Matrigel solution: add 300 μL of Matrigel solution per mL of 0.1 % gelatin.
4 % Paraformaldehyde (PFA) in PBS: add 4 g of PFA to 100 mL of PBS while heating the solution to 65 °C until PFA is completely dissolved. Filter with a 0.20 μm filter before use.
Blocking buffer: 1× PBS supplemented with 5 % FBS and 0.1 % Triton X-100.
Antibody staining buffer: 1× PBS supplemented with 5 % FBS and 0.5 % Triton X-100.
1× PBS (sterile, without magnesium and calcium).
Antibodies: MYH6 clone MF20 (supernatant, Hybridoma Bank), α-Actinin (ascites solution, Sigma), PDGFRA directly labeled with the fluorochrome allophycocyanin (APC) (saline solution, R&D Systems), anti-mouse Alexa 488 or 568 (2 mg/mL, Molecular Probes) (see Note 5).
Optical black 384-well plates.
2× Trypan blue solution.
2.2 Cell Lines
Our preferred human embryonic stem cell line (hESC) H9 for screening carries a MYH6-mCherry reporter, and nuclear PGK1-H2B-GFP reporter [13]. MYH6 is a cardiomyocyte specific gene; by coupling its promoter to a fluorescent reporter we can thus visualize cardiomyocytes being formed. The PGK1-H2B- GFP reporter yields a nuclear GFP signal in every cell driven by the ubiquitous PGK1 promoter, which allows cell counting for toxicity analysis. These reporters thus facilitate high content analysis downstream (see Note 6). Other cell lines are equally suitable, as we have successfully achieved 384-well assays with hESC from the H7 line as well as human induced pluripotent stem cell lines. In case the cell line does not have a reporter, a MYH6 reporter can be inserted easily if needed (we have made lentiviral reporters available on www.addgene.org, plasmids 21228 or 21229, also see Kita-Matsuo et al. [13]) or immunostaining can be implemented for the readout (see Subheading 3.5.2).
Mouse embryonic fibroblasts (MEFs), various commercial sources are available.
2.3 Equipment
Laminar flow cabinet with stereoscope.
Biosafety cabinet.
Automated cell counter, type Countess (Invitrogen) or TC20 (Bio-Rad) or equivalent.
Flow cytometer, type FACSCanto (BD Biosciences) or LSR Fortessa (BD Biosciences) or equivalent.
Liquid handler, type Star (Hamilton Robotics) or equivalent.
Incubator at 37 °C with 5 % CO2.
High content microscope, type InCell (GE Healthcare), Opera (Perkin Elmer), Celigo (Brooks), or equivalent.
3 Methods
We have built hESC/hIPSC differentiation assays to study cardiac differentiation that have several main advantages over classic differentiation protocols (Fig. 1). They are completely serum-free, which focuses the small molecule biology on the differentiation of the cells, rather than on effects of serum components. Secondly, we miniaturized the assay to allow simultaneous screening of thousands of small molecules or small RNAs (see Note 7).
Fig. 1.
Schematic overview of the cardiac differentiation assay. hESC/hIPSC are differentiated with Activin A and Bmp4 in the form of embryoid bodies from day 0 to day 4 to maximize the formation of MESP1+/PDGFRA+ cells. EB are then dissociated to single cells and plated into optical 384-well plates. At this stage small molecules can be added at any desired time or concentration. At day 10 of differentiation, the cells are exposed to T3 to maximize the MYH6 response. At day 18 the plates are processed and imaged on an automated microscope and analyzed automatically
3.1 hESC/hIPSC Culture
Human ESC or IPSC lines can be maintained in the pluripotent state by growing the cells on Matrigel in the presence of MEFs. We describe routine maintenance methods, which we find most suitable for screening purposes. However, the cells can also be maintained feeder-free (see Note 1).
Coat 6-well plates with 1 mL of growth factor reduced Matrigel, overnight at 4 °C. One 6-well plate typically yields enough cells to fill two full 384-well plates in Subheading 3.4.
Plate MEFs in hPSC growth medium at 250,000 cells per well of a 6-well plate, allowing the MEFs to attach overnight before seeding hESC/hIPSC the following day (see Note 8).
Prepare hESC/hIPSC for culture from a frozen vial or from a previous culture. One 6-well plate typically yields enough cells to fill two full 384-well plates in Subheading 3.4.
For recovery of a frozen stock, thaw a frozen vial of cells by plating them in 3 mL of growth medium in the presence of 2 μM Thiazovivin for increased recovery (see Note 9). If splitting from a previous culture, incubate hESC/hIPSC cultures with 1 mg/mL collagenase IV solution for 5 min at 37 °C after removing the growth medium. After the incubation, replace the collagenase IV with growth medium and mechanically dissociate the colonies (by slicing the colonies into small pieces with a 2 mL serological pipet) under a stereoscope and plate at the desired density (see Note 10).
Maintain the hESC/hIPSC in a 37 °C incubator, changing the growth medium daily until cells are confluent. When confluent, cells can be used for freezing, expansion, or differentiation.
3.2 Bulk Differentiation of MESP1/PDGFRA Positive Progenitors
For bulk differentiation of hESC/hIPSC into MESP1+/PDGFRA+ progenitor enriched cultures, we make use of an embryoid body (EB) differentiation step, which is based on a previously described protocol [14]. We pre-passage the hESC/hIPSC onto Matrigel spiked gelatin-coated dishes to facilitate colony removal for EB formation (see Note 11).
Coat 6-well plates with 1 mL per well of 0.1 % gelatin/Matrigel solution. Coating can be done overnight at 37 °C (see Note 12). Once coating is completed, aspirate the coating solution until the plate is completely dry (see Note 13). Two 6-well plates will provide enough cells for one full 384-well plate in Subheading 3.4.
Seed MEFs on plates at 250,000 cells per well in hPSC growth media and allow MEFs to settle overnight before seeding hESC/hIPSCs (see Note 14). Check the following day to ensure attachment of MEFs is sufficient before seeding hESC/hIPSCs. If the MEFs did not attach well, do not continue with these plates, it is best to start over.
Split hESC/hIPSC as described in Subheading 3.1, steps 3 and 4, but now divide the colonies from one confluent well of a 6-well plate to 4 wells of a gelatin/Matrigel coated 6-well plate containing 3 mL of hPSC growth medium per well to allow maintenance of pluripotency.
Allow colonies to grow for 3–4 days, aspirate hPSC growth medium daily and replace with 4 mL of fresh hPSC growth medium.
After 3–4 days the colonies have grown significantly, but should not yet be touching each other. At this stage, remove the hPSC growth medium and incubate the cultures with 1 mL of a 1.5 mg/mL collagenase IV solution for 15 min at 37 °C (see Note 15).
Once the colonies are detached from the plate and are floating in collagenase IV, add 1 mL of hPSC growth medium per well to neutralize the collagenase IV.
Collect the floating colonies and place them into a 15 mL (or 50 mL) conical tube and allow them to settle by gravity pelleting (see Note 16). Rinse the plate with additional hPSC growth medium to collect remaining colonies and add to the conical tube.
Once pelleted, wash the hESC/hIPSC colonies in 2 mL of hPSC growth medium and again allow the colonies to pellet by gravity. Perform an additional wash in 1 mL of hPSC differentiation medium.
Start differentiation for the screening assay (this is day 0 of the assay, the outline of the assay is detailed in Fig. 1) by plating the colonies of one well of a 6-well plate into one well of an ultralow attachment 6-well plate containing 2 mL of hPSC differentiation medium. Add 0.5 ng/mL of Bmp4 to the cultures. Incubate the floating colonies at 37 °C overnight, allowing them to form sphere-like structures, known as embryoid bodies (EB).
At Day 1 of differentiation, aspirate the medium containing the EB from each well with a 5 mL serological pipet and transfer to a 15 mL conical tube. Pellet the EB by gravity. Remove the hPSC differentiation medium and add 2 mL of fresh hPSC differentiation media per well containing 10 ng/mL Bmp4, 3 ng/mL Activin A, and 5 ng/mL bFGF. Return the 2 mL containing the EB to each well of the low attachment plates (see Note 2). Do not continue beyond this point if major cell death is seen or if no EB are formed.
At Day 3, collect EBs by gravity pelleting and refresh hPSC differentiation media again including 10 ng/mL Bmp4, 3 ng/mL Activin A, and 5 ng/mL bFGF (see Note 2).
At Day 4 of differentiation, the EB are ready to be dissociated for plating into the 384-well format (see Subheading 3.3). Alternatively, the EB can be kept for continued differentiation in EB form to test new lots of Activin A and Bmp4 (see Note 17).
3.3 Cell Preparation and Quality Control of Bulk MESP1+/PDGFRA+ Cultures
Before moving ahead with small molecule screening we routinely perform quality control tests on day 4 EB cells.
3.3.1 EB Dissociation and Viability Assessment
At day 4 of differentiation aspirate the medium containing EB and collect 3 wells from a 6-well plate into one 15 mL conical tube by gravity pelleting. Aspirate the hPSC differentiation medium and wash the EB with 5 mL of 1× PBS.
After pelleting again, resuspend the EB in 2 mL of TrypLE per 15 mL conical tube and incubate the 15 mL tubes at 37 °C for 5 min (see Note 18).
Dissociate the EB gently by repetitive resuspending with a 1 mL micropipet tip (see Note 19).
Once dissociated to a single cell suspension (see Note 20), add 6 mL of MEF medium to each 15 mL conical tube containing 2 mL of TrypLE.
Pellet single cells by a 5 min centrifugation at 300 × g.
Aspirate the supernatant and resuspend the cells in 1 mL of hPSC differentiation medium per 15 mL tube.
Remove possible clumps by running the single cell suspension from all 15 mL tubes through a 40 μm cell strainer placed on a 50 mL conical tube. Single cells will flow through and the clumps will remain on the strainer. Discard the strainer at this point. Use the flow through for Subheadings 3.3.2 and 3.4.
Prepare the samples for counting by mixing 10 μL of the cell suspension from Subheading 3.3.1, step 7 with 10 μL of trypan blue. Use 10 μL of this solution to manually or automatically count the cells (see Note 21). Cell viability at this stage is an important quality control checkpoint. If the cells do not have a viability of 90 % or more, we do not proceed with screening (see Note 22).
3.3.2 PDGFRA Expression
At this step the cultures can also be verified for PDGFRA expression as an extra quality control step for the first 4 days of differentiation.
Pellet 105 cells of the cell suspension by centrifugation at 300 × g for 5 min, and resuspend in 100 μL of MEF medium, here used as staining buffer.
Add PDGFRA antibody at 4 μL per 105 cells and incubate for 20 min on ice.
Wash the cells three times in 1× PBS, and analyze the cells on a flow cytometer. Ideally the day 4 cultures should have at least 30 % of PDGFRA+ cells for a reliable screen. Lower yields of PDGFRA+ cells reduce the dynamic range of cardiac induction in the assay.
3.4 Differentiation of MESP1 Enriched Cultures in 384-Well Plates
After checking the viability of the cells and determining incidence of PDGFRA+ cells, the assay is continued in the 384-well format for high throughput screening purposes.
Coat optical 384-well plates with 25 μL of 0.1 % gelatin (this can be done before Subheading 3.3). Dispensing of the gelatin solution can either be done with a 16-channel pipettor (see Note 23) or using a liquid handler for larger scale. Incubate the coated plates for at least 1 h at 37 °C before seeding the cells. Aspirate the gelatin solution using an 8-channel aspirator device.
Seed cells in hPSC differentiation medium including 5 ng/mL of bFGF, at a density of 17,000 cells per well of a 384-well plate, in a total volume of 75 μL per well (see Notes 22 and 24).
Add small molecules at the desired concentration directly into the wells prepared in Subheading 3.4, step 2 (various approaches for compound addition exist) (see Note 25). Compounds can be added any time between day 4 and end of assay at day 18 to probe different time windows of differentiation (see Notes 26–30). siRNAs can also be transfected in at day 4 (see Note 3).
Incubate the plates in a 37 °C incubator until the end of assay (Day 18) (see Note 31).
Depending on the experimental design, compounds can be washed out any day by removing the hPSC differentiation medium and adding 75 μL of fresh hPSC differentiation medium per well.
At day 10 of differentiation, remove the hESC differentiation medium and add 75 μL of SFM including 100 nM of T3 (see Notes 2 and 32) (Figs. 2 and 3). No further media changes are needed until the end of assay at day 18.
By day 14 of differentiation, MYH6 levels should already be elevated and should be visible as a fluorescent signal when using a reporter cell line. We however keep the cultures for 4 more days to allow a further increase of the MYH6 signal, which results in an improved dynamic range for quantification.
At day 18 the assay has completed and the plates are processed for imaging (see Subheading 3.5).
Fig. 2.
Effects of media and serum on the differentiation assay. Differentiation was quantified after the indicated media were used from day 10 of differentiation, the stage when cardiomyocytes form. The use of serum at indicated concentrations suppresses cardiac differentiation compared to StemPro medium. In contrast, use of a range of KOSR containing media or a serum free medium increased differentiation over the level observed StemPro medium
Fig. 3.
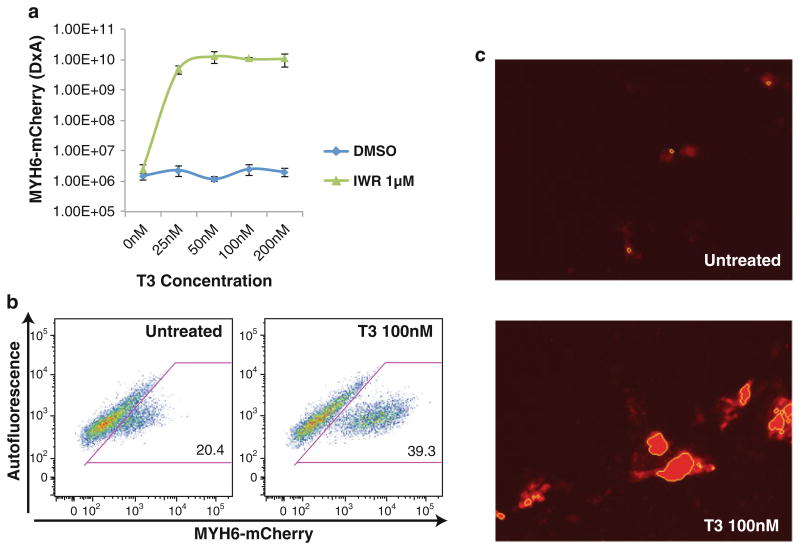
Triiodothyronine (T3) boosts MYH6 expression. The synthetic analog T3 boosts the signal of a MYH6 reporter, but only when cardiomyocyte fate is induced by IWR, as assayed by high content imaging (a). Flow cytometry analysis confirms enhanced MYH6 expression per cell (b). Use of T3 enhances image analysis, as the signal intensity is boosted over tenfold (c), shown using the identical image settings for comparison
3.5 Plate Processing (and Immunostaining if Needed)
When the assay is completed, process the plates for imaging. There are several ways to handle the plates depending on the type of high content microscopes available and depending on whether a reporter line was used or not.
3.5.1 When Reporter Lines Were Used
In the preferred case of a reporter cell line, such as MYH6-mCherry, PGK1-H2B-GFP hESC, which gives a cytoplasmic red and nuclear green fluorescence, the cells can be imaged either live or following fixation. For live imaging, remove the SFM medium from the plate and replace with 25 μL of 1× PBS. Several high content imaging microscopes such as the Celigo can image whole plates very quickly (30–45 min), thus allowing live imaging of numerous plates in a short time frame (see Note 33).
However, when a larger screen of ten plates or more is performed on advanced high content microscopes, remove the SFM medium and fix the cells by adding 25 μL of a 4 % paraformaldehyde (PFA) solution to the wells for 15 min (see Notes 33 and 34). Remove the PFA and wash the cells three times with 1× PBS. After washing, add 50 μL of 1× PBS to the wells. (For longer term storage 50 % glycerol can be used instead of 1× PBS) (see Note 35).
3.5.2 If No Reporter Lines Were Used
Fix the cells as in Subheading 3.5.1.
After fixation, the cell membrane is permeabilized with the detergent Triton X-100 and the cells are blocked to prevent nonspecific antibody binding. To achieve both in one step, add 25 μL of blocking buffer to each well, for 1 h at room temperature, shaking the plates while incubating.
Wash once with 25 μL of 1× PBS.
Incubate the cells with primary antibody, overnight at 4 °C while shaking. Add 25 μL of antibody staining buffer per well, using a 1–100 dilution for the MF20 antibody or 1–500 for α-Actinin.
The next day, wash the wells three times in 1× PBS.
Incubate the cells with secondary antibody and a nuclear stain for 90 min at room temperature while shaking. Stain the cells as follows: add 25 μL of antibody staining buffer per well including 4 μg/mL of secondary antibody labeled with Alexa 488 or 568 and 1 μg/mL of DAPI.
Perform three washes with 25 μL 1× PBS.
Add 50 μL of 1× PBS to the plates (alternatively, 50 % glycerol can be used when storing the plates) (see Note 35).
Plates are now ready to be imaged. Examples of reporter and immunostaining are shown in Fig. 4.
Fig. 4.
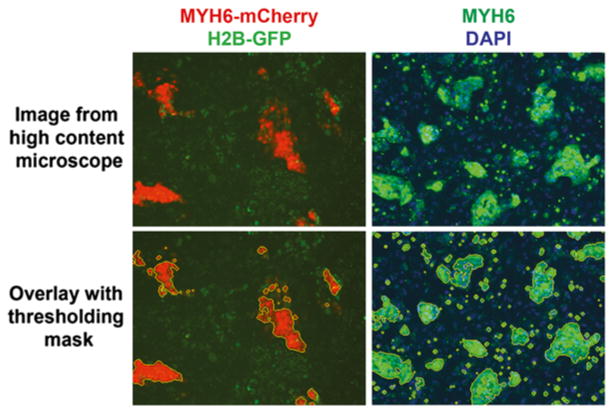
Image examples of the assay readout. Cells engineered to express a fluorescent reporter can be imaged directly (left panels), relying on the red (cardiac MYH6-mCherry) and green (nuclear H2B-GFP) fluorescent signals. Alternatively, immunostaining for MYH6 with an Alexa 488-conjugated secondary antibody can be used to visualize the cardiomyocytes (right panels). Counterstaining with DAPI indicates nuclei. A thresholding algorithm demarcates areas of fluorescent cardiac cells (the mask boundary is outlined in yellow) in both cases
3.6 Imaging and Data Analysis
To quantify cardiac induction, we rely on a high content (HC) imaging approach to image as much of the well surface as possible. The dynamic range of an HC screening approach can be considerably greater than that of a plate reader assay [12, 15]. We here describe the typical flow using an InCell 1000 instrument, yet it is similar for other HC imaging instruments. Images are typically acquired in three color channels (red, green, blue) using filter sets appropriate for the fluorochromes (see Note 33).
Using a 10× objective (numerical aperture = 0.45), 9 fields/well are acquired to maximize imaging of the whole cell surface (see Note 36).
During acquisition, pixels are typically binned at 4 × 4 to reduce the file size (see Note 37).
Once images are collected we run a simple thresholding algorithm to quantify MYH6 expression and nuclear expression. As illustrated in Fig. 4, the MYH6 expression intensity is masked by selecting a certain threshold, and displaying a signal intensity above the background [16] (see Note 38).
After running the algorithm, different parameters can be collected for converting MYH6 expression into a numerical output. We typically report the overall area of the signal captured by the mask to estimate the number of cardiomyocytes formed. We also use the total integrated intensity of the reporter signal within the mask as an estimate of the level of expression per cell. We then multiply both numbers to generate a data output that reflects both the number of positive cells and the expression per cell (typical data are shown in Figs. 2 and 3a). Note that the values only estimate cell number and expression as cardiomyocytes typically grow in tight three-dimensional clusters (see Notes 39 and 40).
Independently, a nuclear count algorithm (also based on thresholding) on the H2B-GFP or DAPI images is run to estimate toxicity of compounds. Reduced levels of nuclei typically indicate toxic effects of the compounds, which aids in discerning toxic compounds from inhibitors when needed.
3.7 Notes
We initially developed this assay by maintaining hESC/hIPSC on Matrigel and MEF feeder layers. While the cells plated on gelatin for differentiation still require MEFs (see Subheading 3.2, steps 1–3), cells for routine maintenance can be grown feeder free on Matrigel using mTeSR medium or TeSR-E8 (Stem Cell Technologies).
These concentrations may vary from source to source (we use proteins from R&D Systems), and batch to batch and cell line to cell line and need to be titrated. Even though we have not seen much Activin A or Bmp4 batch-to-batch variation with our cell lines, each new lot should be carefully titrated. We recommend running an Activin A/Bmp4 array type experiment with twofold doses around our recommended dosages. The EB do not need to be dissociated at day 4 for this purpose (see Note 15).
When we developed the assay, we found that the MYH6-reporter was not bright enough for automated imaging analysis, due to the fact that the cells grew as a single layer of cardio-myocytes in the dish. While we could see weak fluorescence coinciding with contraction by eye, the MYH6-mCherry intensity was not sufficient to quantitatively distinguish signal from background (Fig. 3c top panel). We found that addition of Triiodothyronine (T3), a synthetic thyroid hormone analog selectively boosts the MYH6 signal (Fig. 3a), only when the promoter is active (Fig. 3a) [17]. Flow cytometry clearly demonstrates that T3 causes an increase in the MYH6-mCherry signal, moreover allowing the detection of more cardiomyocytes (Fig. 3b). The impact of T3 for imaging yields a dramatically increased signal to background ratio so that specific signal can be readily detected by a thresholding algorithm (Fig. 3c).
Aside from small molecules, this assay is also suitable for small RNA screening. Various collections of siRNAs are available from different vendors and range from pathway specific to genome wide panels. siRNA transfection per well is performed by growing the cells in 65 μL of hESC differentiation medium, supplemented with 0.1 μL of RNAiMax, siRNAs at a desired concentration in water up to 5 μL and Opti-MEM added to complement the siRNAs up to 10 μL. After 24 h, the medium can be replaced with 75 μL of hPSC differentiation medium.
Other vendors provide similar antibodies that may replace the ones indicated. Careful titrations would be required when alternate options are used as the concentrations listed here are based on the antibodies listed.
The use of a reporter line has multiple benefits for the high content screening process: (a) assay development is facilitated as reporter expression can be followed in real time. (b) Assay controls are immediately visible before processing the plates as described. (c) Antibody-based read outs are costly and can be prohibitive when running larger screens.
hESC/hIPSC cultures in our hands are less consistent than mouse PSC, and therefore similar assays for mouse PSC have historically allowed for larger scale screens than the hESC/hIPSC assays. Our experience suggests that hits identified in either assay can be translated to the other. To perform a mouse assay, EB from a Myh6-GFP reporter line are formed in SFM by plating 50,000 cells per mL in non-coated dishes. The assay similarly relies on the exposure to Activin A and Bmp4 from day 2 to day 4, after which the cells are plated into 384 wells at 8,000 cells per well. IWR then is a key switch to turn on cardiac fate at day 5, with cardiac induction seen by GFP expression at day 7.
Alternatively MEFs can be seeded simultaneously with hESC/hIPSC, but pre-plating of MEFs is recommended.
We typically freeze one confluent well of a 6-well plate into one 2 mL cryovial. Since recovery after thawing is never 100 %, we thaw one vial to one well of a 6-well plate.
We typically passage 1–6, meaning one confluent well of a 6-well plate is sufficient for 6 wells of a 6-well plate.
EB formation from intact colonies is much more efficient than from pieces of cut up colonies. To facilitate removal of colonies, we therefore switch Matrigel for a solution of 0.1 % gelatin and a low amount of Matrigel to coat plates, providing just enough attachment to maintain pluripotency.
Alternatively, plates can be generated the same day at least 2 h before seeding MEFs, again placing the coated plate at 37 °C. Overnight coating is however recommended.
To facilitate drying of the plates, we found that leaving plates open in a biosafety cabinet for about 1 h is sufficient. Drying is important, as we noted that hESC/hIPSC colonies tend to peel off after 48 h if the plates were not fully dried.
We use MEFs to preserve the pluripotency and compact nature of the PSC colonies for differentiation. We tend to get better differentiation results compared to hESC/hIPSC maintained on Matrigel in MEF conditioned hPSC growth medium or mTeSr.
Colonies should lift off easily. Otherwise the plate can be tapped gently or agitated to facilitate colony lifting. Manual dislodging may also help. We do not recommend incubation for longer times as we have found that longer exposure affects survival of EB. If needed, slightly increase collagenase IV concentration, but avoid concentrations above 2 mg/mL.
Gravity pelleting of colonies or EBs selectively pellets viable colonies/EB and avoids dead cells, which would be present if colonies were collected by centrifugation. The media containing the dead cells should however be aspirated immediately after the colonies or EB have settled at the bottom of the tube. Prolonged times of gravity pelleting will also allow dead cells to pellet.
To verify that EB differentiation progressed normally from day 0 to 4 or to optimize day 1–4, a differentiation control can be used. Day 4 EB are plated on 0.1 % gelatin coated dishes in hPSC differentiation medium in the presence of 1 μM IWR and 5 ng/mL bFGF. Cardiac induction can be monitored by beating, reporter expression, immunostaining, RT-qPCR for cardiac markers such as TNNT2 or MYH6 or by flow cytometry with a SIRPA antibody [6, 18].
We prefer to use 15 mL conical Falcon tubes, but 50 mL conical tubes are also suitable. Note that the volume should not be increased above 5 mL (it is important to maintain the EB to TrypLE ratio), as this will affect the efficiency of dissociation with a 1 mL micropipet tip.
We do not recommend the use of serological pipets for dissociation, as the opening is too wide for efficient disruption of EB to single cells.
EB are large clumps that are clearly discernable by eye. As dissociation takes place, these clumps will dissolve and should disappear. Manual resuspension is essential to remove final clumps. If clumps or strands of cells are observed after this process, do not continue dissociation, but rather continue the process. Chunks will be removed subsequently using the cell strainer.
For cell count consistency between screens, we use an automated cell counter such as the Invitrogen Countess or the Bio-Rad TC20. Both count cells and determine their viability using trypan blue. Alternatively a hemocytometer can be used.
Cell preparations with lower viability may still differentiate properly, but in our hands, under these conditions the risk of failure is too high to warrant continuation of the screening efforts. We do use these lower viability preparations for smaller secondary assays.
We use automatic 16-channel pipets (such as the Matrix series from Thermo Scientific) as they allow repeated accurate dispensing of volumes down to 2 μL. Manual alternatives exist, but use of such pipets will increase row-to-row or column-to-column variation and we do not recommend these for screening purposes.
Cell number can be increased to ensure proper differentiation, and the number should be re-titrated when developing this assay, as counting can vary from counter to counter. If viability is lower than 90 % it is possible to increase cell number per well, but this is only recommended when running small-scale hit confirmation experiments.
For compound addition, we use an acoustical spotter (Echo 550, Labcyte Inc), which dispenses nanoliter volumes of compound directly into the wells of a 384-well plate, which already contain cells and medium in most cases. An alternative for nanoliter transfers are pintools, which also directly transfer small molecules into the wells containing medium and cells. Both the acoustic spotter and pintool can also be used to spot the compounds in the gelatin coated 384-well plates before the cells and medium are added. Pintools and acoustic spotters are less accessible and an alternative approach using liquid split as hand-lers may be used to add compounds diluted in hESC differentiation medium, transferring the compounds in a volume of 2–5 μL to the cells. To achieve effective working concentrations, the compound libraries should be sub-diluted into hPSC differentiation medium to a 20–35× concentrated working solution.
Allow cells to settle to the bottom of the plate by gravity at room temperature. Centrifugation or incubation at 37 °C will give a less uniform distribution, and an even distribution is essential for optimal cardiac differentiation down the line.
The assay can be run in agonist mode to identify inducers of cardiac fate or in antagonist mode to identify molecules that block cardiac differentiation. In agonist mode, day 4 EB cells are not induced to form cardiomyocytes, and the added small molecules or siRNAs are expected to promote cardiac differentiation. In antagonist mode, however, the day 4 cells are exposed to 1 μM of IWR, which will induce cardiac differentiation efficiently. Small molecules or siRNAs added can then be used to block cardiac differentiation to probe the signaling or genetic cascades involved.
Positive control compounds to induce cardiac differentiation at day 4 include Wnt inhibitors [6] and additional treatment with the Nodal/TGFβ inhibitor SB-431542 [19].
-
The choice of libraries is an important upfront decision. We have screened collections of relatively uncharacterized molecules selected for chemical diversity (for example the Chembridge DiverSet collection). Such libraries are frequently used in target-based screens, which are designed to identify hits that elicit a particular biochemical activity (e.g., enzyme inhibition), often through a constrained molecular mechanism of action, against the target. These libraries are typically assembled based on their representation of chemical diversity space, comprising structures that have been deemed likely to engage the target of interest. However, since relatively little is known about the biological activities of these molecules, identification of the biological mechanism of action and the actual target typically requires a vast amount of work. Nevertheless, such collections are more likely to identify novel mechanisms and in our hands resulted in a completely novel TGFβ inhibitor [7, 9].
An alternative approach to gain biological insight is to screen focused collections of small molecules that selectively engage known sets of cellular proteins. We typically use StemSelect and Inhibitor Select collections from EMD/Millipore or the Lopac 1280 collection from Sigma. Although small molecules are rarely selective for a unique protein, following the known protein targets of screen hits in this case has proven an effective strategy for identifying cellular processes that control complex biological phenomena. For example, the use of small molecule pathway modulators facilitated the discovery of biological mechanisms that drive IPSC generation of cardiac differentiation [6, 20].
The number of replicates necessary to discern hits should be determined during assay development, and a useful discussion is in the reference by Zhang et al. [21]. We screen in triplicate to ascertain hits since dynamic range of the cardiac differentiation protocol is typically insufficient to screen without replicates.
Place extra water pans in the incubator to ensure a well-humidified environment to limit evaporation of media from the wells.
StemPro 34 supports differentiation of hESC/hIPSC to cardio-myocytes, but our experience is that the cardiomyocytes are less viable. Evaluating different media, we found that use of certain serum-free options, including Knock Out Serum replacer or B27/N2 supplement based media, yield healthy cardiomyo-cytes. Serum-free media also prevents fibroblast overgrowth, thus enhancing signal to background levels (Fig. 2).
Currently, there are many different high content imagers that can be used for reading out this assay. The relatively inexpensive Celigo platform images at lower resolution enabling quick whole well imaging with sufficient quality for image analysis. Other options include the InCell series from GE Healthcare or the Opera/Operetta series from Perkin Elmer, allowing higher resolution images at different magnifications. For quantifying the cytoplasmic fluorescent signal as described in this protocol, the Celigo, InCell, and Opera platforms provide similar signal to background and dynamic range.
Longer fixations are not recommended to avoid loss of signal from fluorescent proteins and increased background.
Alternatively 50 μL of 50 % glycerol in PBS can be added to the plates to preserve the fluorescent signal for several weeks.
Partial field or higher magnification acquisition works, but we prefer to capture as much of the well as possible as cardiac differentiation typically occurs in an unpredictable and unevenly distributed pattern.
This reduces resolution of the images, and thus overall quantity of data, and consequently speeds up processing time. It does not affect quantification of the cytoplasmic stain. However, binning is not appropriate for higher resolution imaging.
We use a custom-built algorithm in the imaging package Cyteseer (Vala Sciences Inc.) to generate masks. The algorithms permit manual determination of the threshold to be used (based on principles described by Bushway et al. [16]). Most current HCS instruments include an image analysis package that can run similar thresholding operations.
Secondary assays using proper quantification methods such as flow cytometry should be used to quantify expression level and incidence of positive cells.
An alternative output to estimate cardiomyocyte yield is the count of MYH6 positive objects. We typically do not use this metric, instead prefer secondary assays that directly quantify incidence (see Note 39).
Acknowledgments
This work was supported by grants from the California Institute for Regenerative Medicine (CIRM) (RC00132) and NIH (HL113601, HL108176, and AG043917) to M.M. E.W. was supported by American Heart Association and CIRM training grants.
References
- 1.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Mercola M, Colas A, Willems E. Induced pluripotent stem cells in cardiovascular drug discovery. Circ Res. 2013;112:534–548. doi: 10.1161/CIRCRESAHA.111.250266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 4.Burridge PW, et al. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garbern JC, Lee RT. Cardiac stem cell therapy and the promise of heart regeneration. Cell Stem Cell. 2013;12:689–698. doi: 10.1016/j.stem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willems E, et al. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willems E, et al. Small molecule-mediated TGF-β type II receptor degradation promotes cardiomyogenesis in embryonic stem cells. Cell Stem Cell. 2012;11:242–252. doi: 10.1016/j.stem.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanier M, et al. Wnt inhibition correlates with human embryonic stem cell cardio-myogenesis: a structure–activity relationship study based on inhibitors for the Wnt response. J Med Chem. 2012;55:697–708. doi: 10.1021/jm2010223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schade D, et al. Synthesis and SAR of b-annulated 1,4-dihydropyridines define car-diomyogenic compounds as novel inhibitors of TGFβ signaling. J Med Chem. 2012;55:9946–9957. doi: 10.1021/jm301144g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borowiak M, et al. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, et al. A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol. 2009;5:258–265. doi: 10.1038/nchembio.154. [DOI] [PubMed] [Google Scholar]
- 12.Willems E, Bushway PJ, Mercola M. Natural and synthetic regulators of embryonic stem cell cardiogenesis. Pediatr Cardiol. 2009;30:635–642. doi: 10.1007/s00246-009-9409-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kita-Matsuo H, et al. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One. 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR + embr yonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 15.Bushway PJ, Mercola M, Price JH. A comparative analysis of standard microtiter plate reading versus imaging in cellular assays. Assay Drug Dev Technol. 2008;6:557–567. doi: 10.1089/adt.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bushway P, Mercola M. High-throughput screening for modulators of stem cell differentiation. Methods Enzymol. 2006;414:300–316. doi: 10.1016/S0076-6879(06)14017-3. [DOI] [PubMed] [Google Scholar]
- 17.Gustafson TA, et al. Thyroid hormone regulates expression of a transfected alpha-myosin heavy-chain fusion gene in fetal heart cells. Proc Natl Acad Sci U S A. 1987;84:3122–3126. doi: 10.1073/pnas.84.10.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubois NC, et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai W, et al. Coordinate Nodal and BMP inhibition directs Baf60c-dependent cardiomyocyte commitment. Genes Dev. 2013;27:2332–2344. doi: 10.1101/gad.225144.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Rana TM. A kinase inhibitor screen identifies small-molecule enhancers of reprogramming and iPS cell generation. Nat Commun. 2012;3:1085. doi: 10.1038/ncomms2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Chung T, Oldenburg K. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]