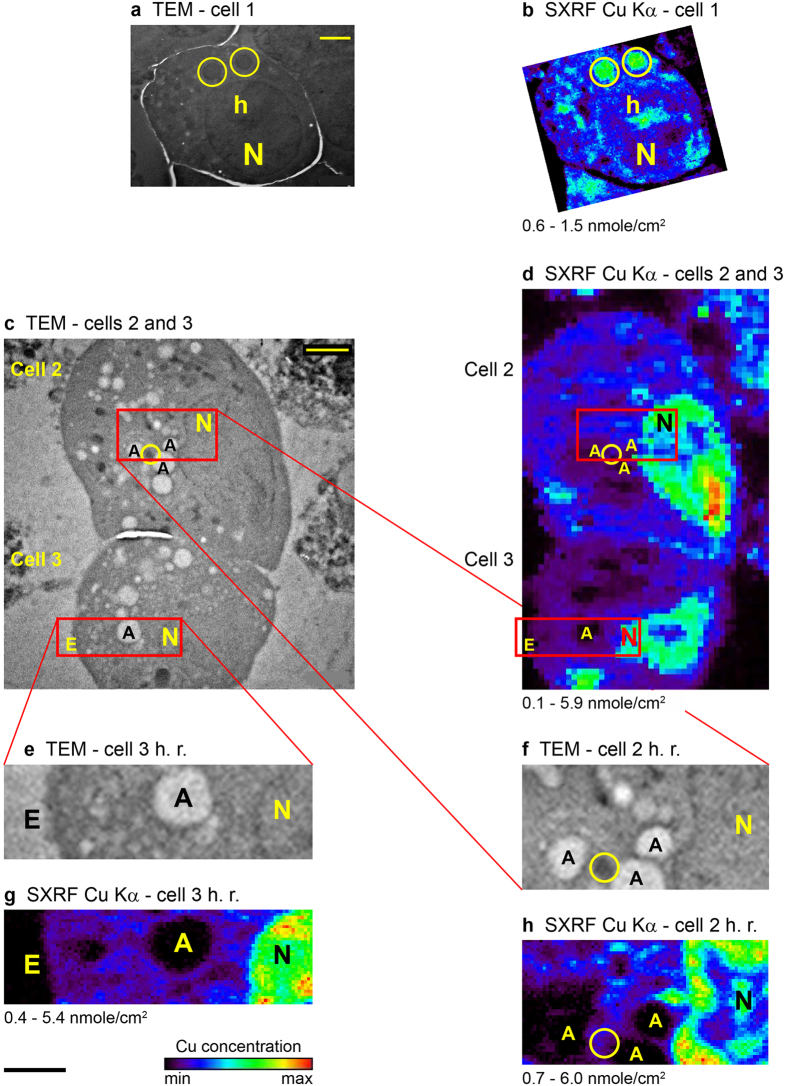
Figure 1. Complementary TEM and SXRF micro/nanoprobe Cu Kα distribution maps of MIN6 cells 1–3.
The combined information from the two imaging techniques gives a detailed ultrastructural and chemical picture of the subcellular environment at the single organelle level. (a) TEM map of cell 1. This cell was grown in a medium with no Cd. It serves as a reference for the sample preparation and imaging procedure we used. Scale bar = 2 μm. (b) The corresponding SXRF microprobe map to a. Scan step size = 0.1 μm, dwell time = 1 s. Same scale as a. The two Au nanoparticles that are seen in the map, as red pixels in the upper and lower left parts, are off the reported concentration range. (c) TEM image of cells 2 and 3. These cells were grown in a medium supplemented with 1 μmole/l CdCl2. Red rectangles outline the areas imaged with the SXRF nanoprobe at high-resolution. Scale bar = 2 μm. (d) The SXRF microprobe map corresponding to c. Scan step size = 0.25 μm, dwell time = 1 s. Same scale as c. (e–f) Enlargements of the high-resolution (h. r.) scan areas in the TEM image of cells 3 and 2 (c), respectively. (g–h) The SXRF nanoprobe maps corresponding to e–f. Scan step size = 0.05 μm, dwell time = 5 s. One μm scale bar (below g) applies to (e–h). Elemental spatial density scale bar (below g) applies to (b,d,g–h) and the measured range, in units of nmole/cm2, is indicated below each map. Contrast of the SXRF images (b,d,g–h) has been enhanced. A – autophagosome-like feature, E – edge of the cell, h – heterochromatin, N – nucleus, circles outline mitochondria. Additional concentration information, including uncertainties, appears in Supplementary Table S2.