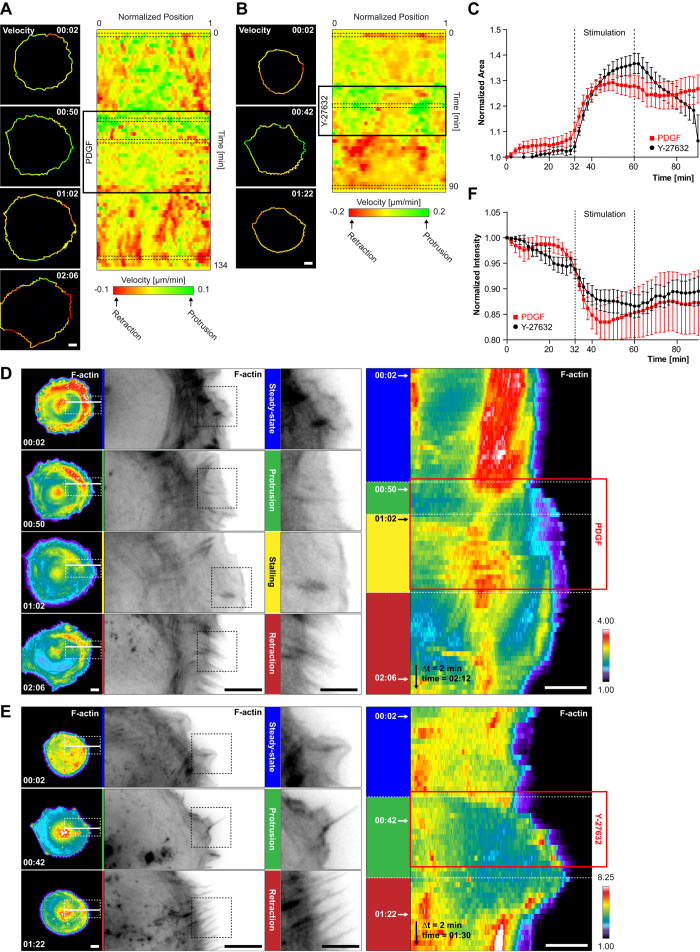
Figure 1. PDGF/Y-27632 pulse-induced edge and F-actin dynamics.
(A,B) Quantification of edge dynamics upon PDGF (A) or Y-27632 (B) stimulation. The ADAPT Image J plugin30 was used to extract edge dynamics and velocities using the Lifeact-mCherry signal. Left panel: cell outlines display color-coded protrusion/retraction velocities (Time scale: hours:minutes). Right panel: velocity maps along the entire cell edge at normalized positions. Black dashed contours indicate specific cell outlines shown in the left panel. Color-code according to the scale bar. (C) Cell area dynamics in response to PDGF or Y-27632. Cell area was measured using ADAPT, and normalized to t = 0′. Average area ± s.e.m (PDGF: n = 12 cells, Y-27632: n = 13 cells). (D,E) F-actin dynamics during protrusion/retraction in response to PDGF (D) or Y-27632 (E). F-actin signals are color-coded for signal intensity or shown in inverted black and white (ibw) contrast. Color-code according to the scale bar. Left panel: F-actin signal (color-coded) of representative whole cells (left), as well as magnified insets (successively corresponding to the white and black dashed boxes). White solid lines represent ROIs used for kymograph analysis. Right panel: Kymograph analysis of F-actin dynamics during the PDGF/Y-27632 pulse. (F) Quantification of F-actin fluctuations. Whole cell lifeact-mCherry fluorescence intensity was averaged in response to a PDGF/Y-27632 pulse. Intensities were normalized to t = 0′. Average intensity ± s.e.m (PDGF: n = 5 cells; Y-27632: n = 5 cells) Scale bars: (A,B,D,E) 10 μm.