Fig. 4.
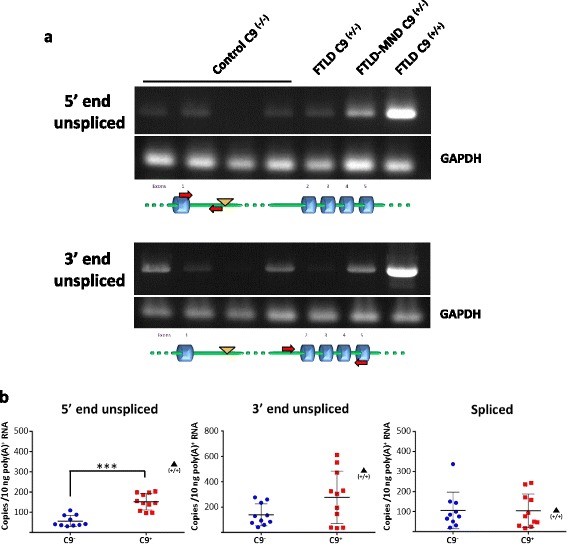
Intron 1 retention in C9orf72 transcripts in the brain of C9orf72 expansion carriers. C9orf72 transcripts were analyzed in the frontal cortex from frontotemporal lobar degeneration (FTLD) or frontotemporal lobar degeneration with motor neuron disease (FTLD-MND) cases with confirmed C9orf72 hexanucleotide expansions and from control individuals. a RNA was analyzed by RT-PCR as in Fig. 1. GAPDH was used as a loading control. The level of C9orf72 transcripts unspliced at the 5’ and 3’ ends in an FTLD case homozygous for the C9orf72 G4C2 repeat expansion was markedly higher than in heterozygous cases (C9(+/+), far right lane). b Quantitative analysis of intron retention by real-time PCR. Levels of C9orf72 transcripts spliced or unspliced at the 5’ and 3’ end of intron 1 were determined by real-time qRT-PCR in heterozygous expansion carriers (n = 11) and control cases (n = 10). Data are shown as means ± SEM. Each data point represents an individual case, ***P < 0.001, Mann–Whitney U test. C9−, controls; C9+, expansion carriers. ▲(+/+) indicates the values for the single homozygous case analyzed
