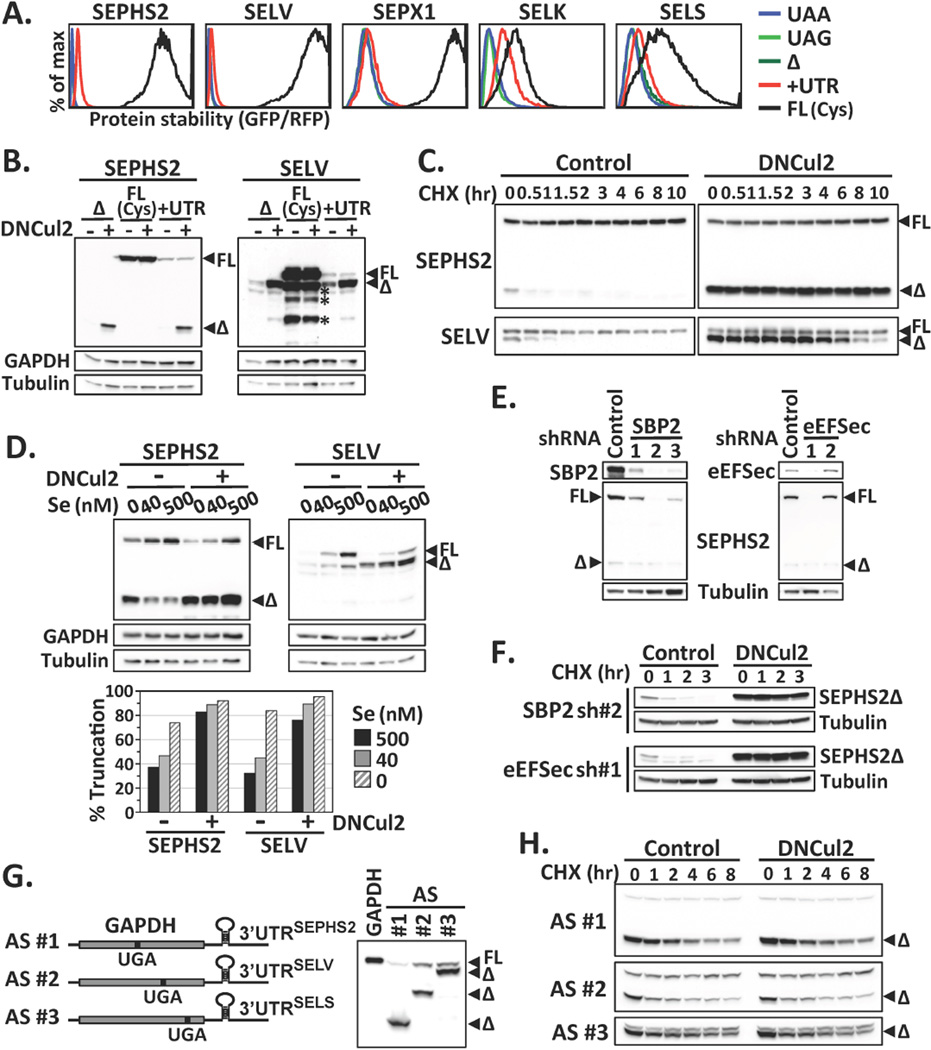
Figure 2.
Failures in Sec incorporation and selenoprotein degradation. (A) Protein stability comparison among various forms of selenoproteins by GPS. (B) Western blot analysis of SEPHS2 or SELV mutants. Asterisks indicate degradation products from full-length SELV. (C) The stability of SEPHS2 or SELV proteins expressed from the UTR construct was subjected to cycloheximide (CHX)-chase analysis. (D) Cells expressing SEPHS2 or SELV from the UTR construct were cultured in serum-free medium supplied with graded increase of extracellular sodium selenite, with or without DNCul2 treatment, and analyzed by Western blotting. The percentage of truncated selenoprotein is shown below. (E) Cells expressing SEPHS2 from the UTR construct were treated with shRNAs for SBP2 or eEFSec and then analyzed by Western blotting. (F) The protein stability of SEPHS2 in cells treated with shRNAs for SBP2 or eEFSec was analyzed by CHX-chase. (G) A schematic representation of GAPDH artificial selenoprotein (AS) transcripts. Cells expressing wild-type GAPDH or AS were analyzed by Western blotting. The introduced in-frame UGA codon terminated GAPDH at the amino acid positions 152, 247 and 301. (H) CHX-chase analysis of GAPDH expressed from AS transcripts in (G).