Extended Data Figure 2. Generating the mouse strains.
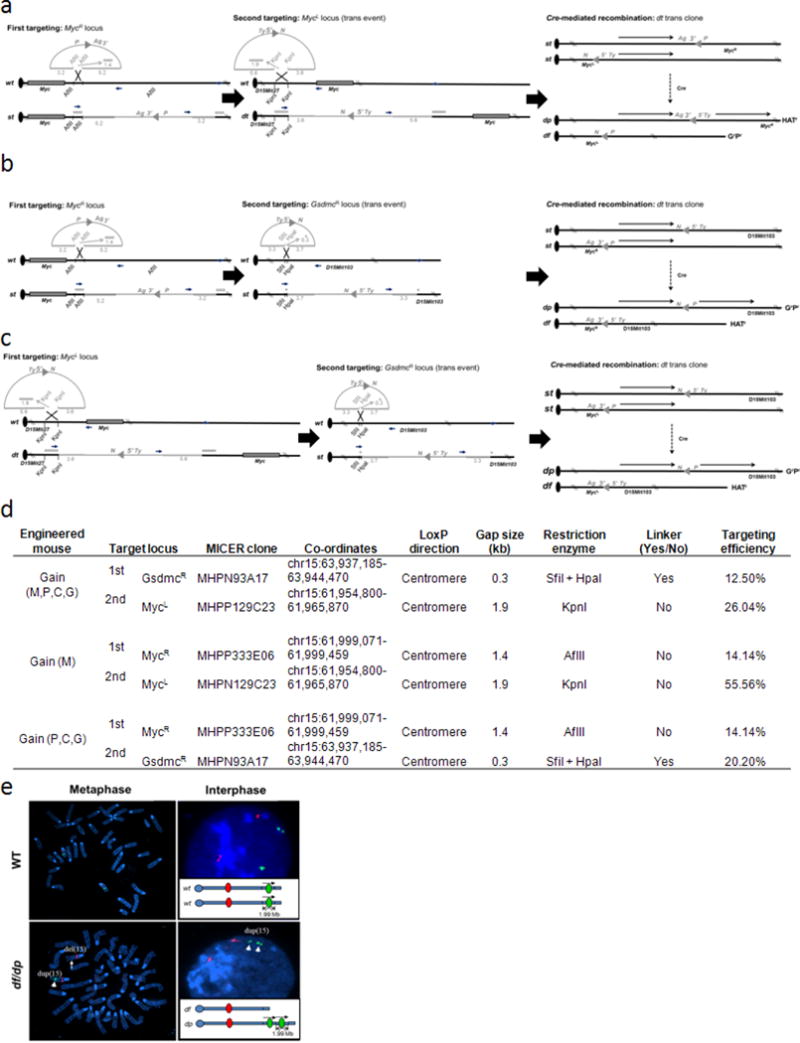
a–c, Chromosome engineering of Hprt-deficient mouse AB2.2 embryonic stem cells was used to develop mouse strains containing an extra copy of Myc (gain(Myc)) (a), Pvt1/Ccdc26/Gsdmc (gain(Pvt1,Ccdc26,Gsdmc)) (b) and Myc/Pvt1/Ccdc26/ Gsdmc (gain(Myc,Pvt1,Ccdc26,Gsdmc)) (c). Gene targeting was performed in AB2.2 with targeting vectors obtained from the Mutagenic Insertion and Chromosome Engineering Resource (MICER) that were modified to facilitate detection of correctly targeted clones by PCR. These electroporated embryonic stem cells were cultured in G418 (G, 180 µg ml−1) or puromycine (P, 3 µg ml−1) for 7–10 days. Correctly targeted clones were identified by PCR analysis. Double-targeted embryonic stem cells were electroporated with the transient Cre-recombinase expression vector pOG231. After subsequent selection of recombinants by using hypoxanthine aminopterin thymidime (H) media for 7 days and then recovery of recombinants by using hypoxanthines and thymidine media for 2 days, the HRGRPR clones containing one mouse chromosome 15 with the targeted region duplication (dp) and the other mouse chromosome 15 with the targeted region deletion (df) were identified. d, Summary of gene targeting of mouse genome at the MycL, MycR and GsdmcR loci. e, FISH analysis of gain/loss Myc,Pvt1,Ccdc26,Gsdmc embryonic stem cells with balanced allele for the Myc/Pvt1/Ccdc26/Gsdmc region. Metaphase and interphase preparations from the engineered cells were probed with BAC clone specific for chromosome 15 and located outside (RP23-18H8, red) and within (RP24-78D24, green) the engineered region. The alleles containing the deletion and the duplication are marked.
