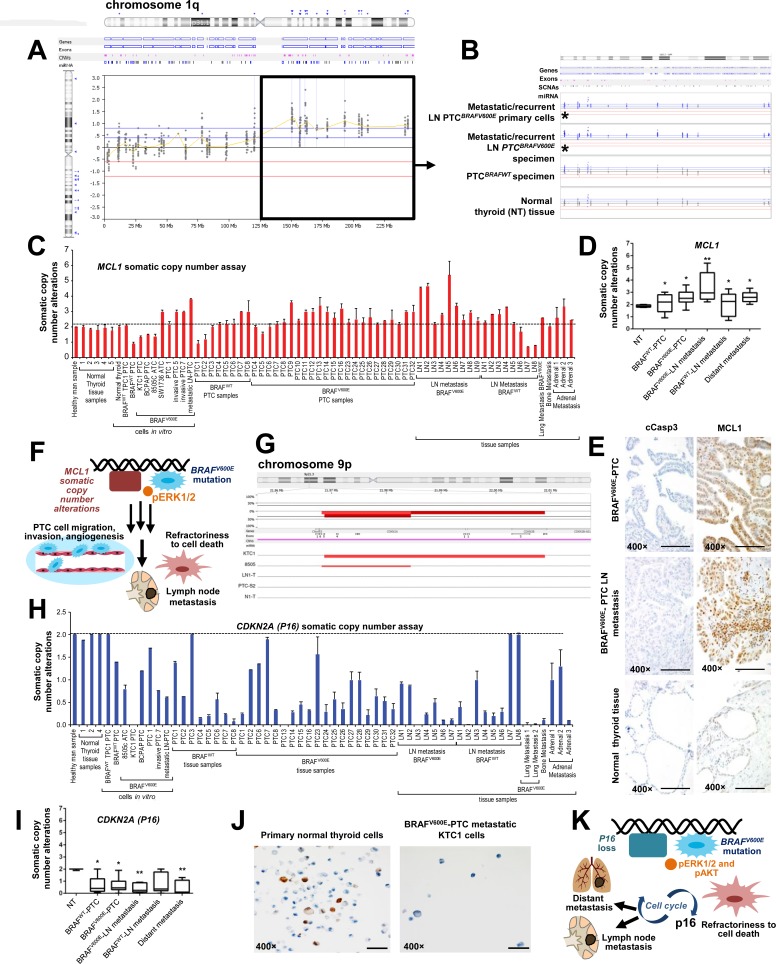
Figure 4. Analysis of MCL1 somatic copy number gain (chromosome 1q) and loss of P16 (chromosome 9p) in non-metastatic or metastatic papillary thyroid carcinoma (PTC) samples.
A. Graphical representation of the log2 ratio of sequence coverage in tumor versus reference for fragments sequenced using a targeted exome sequencing strategy. BRAFV600E-PTC primary cells or patient specimens harboring the BRAFV600E mutation were compared with PTC or NT samples with BRAFWT. This analysis revealed 1q somatic copy number alterations (SCNAs) (i.e. copy number gain/amplifications) in metastatic/recurrent neck or mediastinal lymph nodes (LN). B. Detailed view of sequenced exons in 1q, including the MCL1 gene highlighted by asterisks, in metastatic/recurrent mediastinal LN (lymph node) BRAFV600E-PTC primary cells and patient specimens harboring the BRAFV600E mutation compared with PTC or NT samples with BRAFWT. Probes that were colored or shaded blue or marked by blue triangles were called as gained by the analysis software. C. MCL1 somatic copy number analysis normalized in 31 samples which included PTC and normal thyroid samples, and some established primary PTC or NT cell cultures derived from same patients' cohort, from independent patients. Histogram shows MCL1 copy number assay results in: 1 DNA control sample from healthy man; 1 primary NT (normal thyroid) cell culture; 3 primary PTC cell cultures (2 with BRAFWT/V600E and 1 with BRAFWT), 1 mediastinal LN metastatic BRAFV600E-PTC cell culture; 5 spontaneously immortalized cell lines established from patients with BRAFV600E (i.e., KTC1 and BCPAP) or BRAFWT(TPC1) PTC, or BRAFV600E-positive anaplastic thyroid cancer (ATC) (i.e. 8505c and SW1736); 5 NT (normal thyroid) tissue samples, 31 PTC (23 BRAFV600E-PTC and 8 BRAFWT-PTC), 17 LN metastatic PTC (9 BRAFV600E-LN and 8 BRAFWT-LN) and 5 distant metastatic samples from patients with PTC (1 from lungs, 1 from bone and 3 from adrenal glands). Results were normalized against two separate reference, housekeeping (reference) genes: GAPDH and RNAase-P. These data represent the average ± standard error mean (error bars) of 2–3 independent replicate measurements. D. Box plot analysis using: 5 NT (normal thyroid) tissue samples, 31 PTC (23 BRAFV600E-PTC and 8 BRAFWT-PTC), 17 LN metastatic PTC (9 BRAFV600E-LN and 8 BRAFWT-LN) and 5 distant metastatic samples from patients with PTC (1 from lungs, 1 from bone and 3 from adrenal glands). These data represent the average ± standard deviation of 2–3 independent replicate measurements (*p < 0.05, **p < 0.01, ***p < 0.01, one-way ANOVA test). E. Immunohistochemistry shows strong and diffuse MCL1 protein expression in the cytosol and stippled nuclear staining in all neck LN metastatic PTC samples harboring BRAFV600E (n=3) (scoring, 3+) and primary PTC with BRAFV600E (n=4) (scoring, 3+), whereas NT samples (n=3) (scoring, 1+) showed focal and weak nuclear MCL1 localization. Primary BRAFV600E-PTC (scoring, 0) or LN metastatic BRAFV600E-PTC (scoring, 0) samples did not show significant change in cytoplasmic cleaved caspase 3 (cCasp3) protein levels compared with NT samples (scoring, 0). Protein expression was assessed semi-quantitatively using the following scoring method: 0 (negative), 1+ (1–10% positive cells), 2+ (11–50% positive cells), and 3+ (more than 50% positive cells). All scale bars are=400 μ. F. Proposed mechanisms of LN metastatic PTC spreading from primary PTC harboring the BRAFV600E mutation along with MCL1 copy number gain/amplification. G. Graphical representation of next generation sequencing, targeted-exome sequencing results showing loss of P16 (marked by red bars) in the chromosome 9p in PTC (i.e. heterozygous BRAFWT/V600E-positive human KTC1 PTC-derived cells with P16 homozygous loss) or in ATC samples with distant metastasis (i.e. homozygous BRAFV600E-positive 8505c ATC cells) compared with metastatic/recurrent LN BRAFV600E-PTC sample (LN1-T) or derived primary cells (e.g. LN0-PTC), BRAFV600E-PTC sample (e.g. PTC-7), BRAFWT-PTC sample (e.g. PTC-S2), or NT sample with BRAFWT (e.g. N1-T) which show neutral copy number without loss of P16. The top red bar shows the aggregate SCNAs for all samples. H. P16 somatic copy number alteration analysis normalized in: 1 DNA control sample from healthy man; 3 primary PTC cell cultures (2 with BRAFWT/V600E and 1 with BRAFWT), 1 mediastinal LN metastatic BRAFV600E-PTC cell culture; 4 spontaneously immortalized cell lines established from patients with BRAFV600E (i.e., KTC1 and BCPAP) or BRAFWT(TPC1) PTC, or BRAFV600E-positive anaplastic thyroid cancer (ATC) (i.e. 8505c); 4 NT (normal thyroid) tissue samples, 27 PTC (19 BRAFV600E-PTC and 8 BRAFWT-PTC), 15 LN metastatic PTC (7 BRAFV600E-LN and 8 BRAFWT-LN), 6 distant metastatic samples from patients with PTC (2 from lungs, 1 from bone and 3 from adrenal glands). Results were normalized against two separate reference, housekeeping (reference) genes: GAPDH and RNAase-P. Histogram shows P16 copy number assay. These data represent the average ± standard error mean (error bars) of 2–3 independent replicate measurements. I. Box plot analysis using: 4 NT (normal thyroid) tissue samples, 27 PTC (19 BRAFV600E-PTC and 8 BRAFWT-PTC), 15 LN metastatic PTC (7 BRAFV600E-LN and 8 BRAFWT-LN), 6 distant metastatic samples from patients with PTC (2 from lungs, 1 from bone and 3 from adrenal glands). These data represent the average ± standard deviation of 2–3 independent replicate measurements (*p < 0.05, **p < 0.01, ***p < 0.01, one-way ANOVA test). J. Immunocytochemistry shows loss of P16 protein expression in human KTC1 PTC-derived cells (BRAFWT/V600E spontaneously immortalized metastatic PTC cells) compared with primary NT cells which show P16 protein focal staining in the nuclei. All scale bars are=100 μ. K. Proposed mechanisms of metastatic spreading from primary PTC harboring the BRAFV600E mutation along with loss of P16.