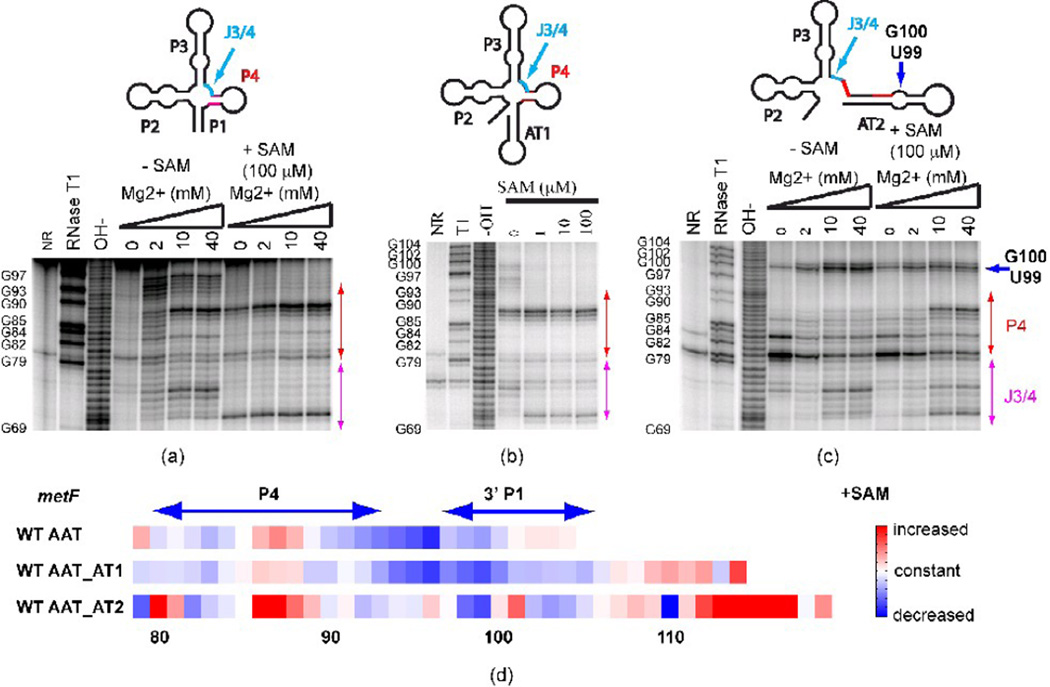
Figure 2. In-line probing of secondary structure in metF SAM-I riboswitch-containing transcripts with varying 3’ truncation points.
(a) Truncation at the aptamer, (b) Truncation at the “switch point”, at which residues comprising AT1 have been transcribed (c) Truncation at the point of complete transcription of AT2 helix. Red arrows: change of cleavage pattern in the P4 helix. Blue arrow: the increased cleavage is likely to be due to the GU bulge in the alternative AT helix model. J3/4 region is indicated as the arrow in cyan. (d) Quantification of the effect of SAM on in-line probing experiments (40 mM Mg2+ lanes) for three metF SAM-I riboswitch RNA constructs starting from nucleotide 79. The color scale is shown on the right—red means increased cleavage rate, white denotes no change and blue indicates decreased cleavage rate in the presence of SAM. WT AAT—truncation at the aptamer; WT AAT_AT1— truncation at the “switch point”, at which residues comprising AT1 have been transcribed; WT AAT_AT2—truncation at the point of complete transcription of AT2 helix. Note that RNAse T1 cleavage bands for G100, G102, and G104 are not resolved from the full-length uncleaved band. Altogether, these results indicate that P1 helix and OFF state formation are inhibited in long metF SAM-I riboswitch transcripts by the formation of the AT2 helix.