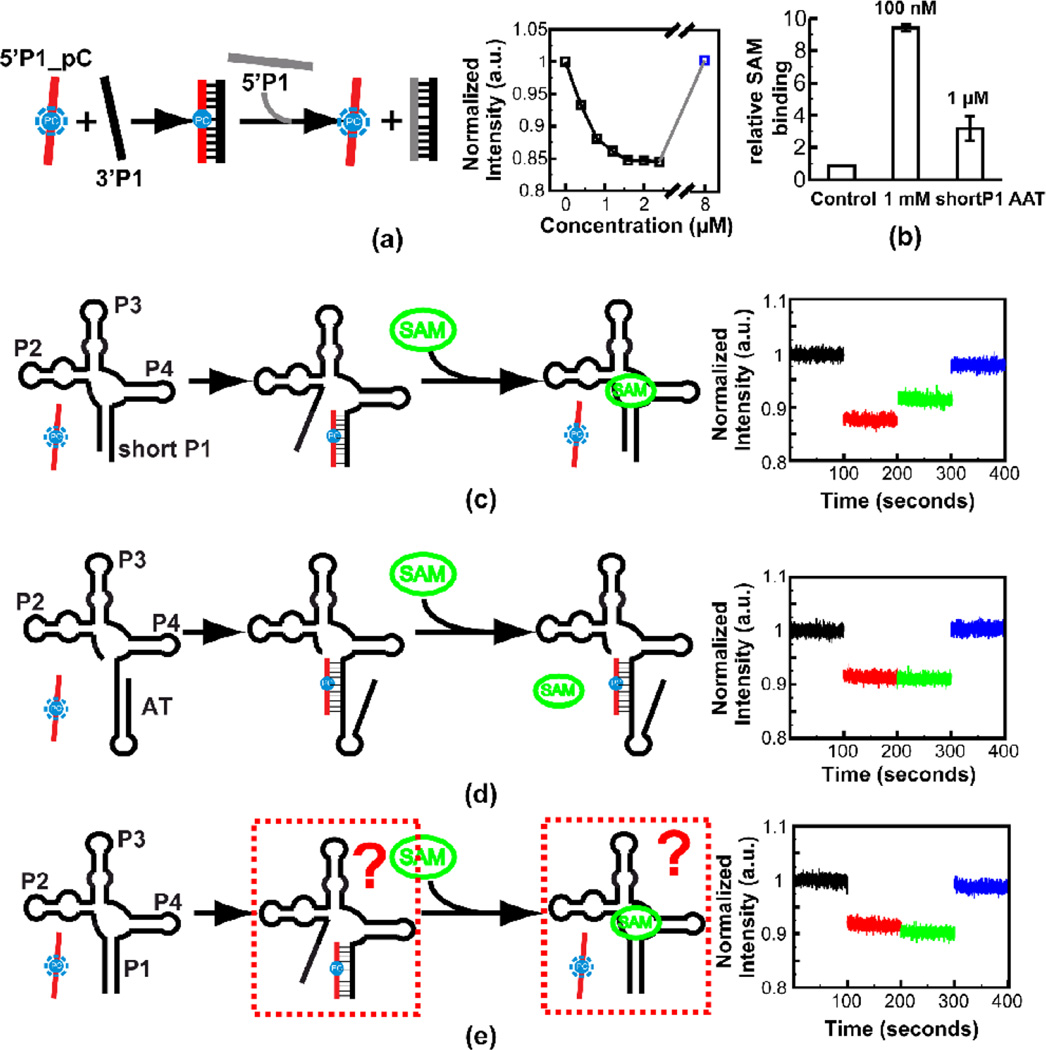
Figure 7. Steady state fluorescence experiment on the metF SAM-I riboswitch RNA constructs.
(a) Titration of 3’ strand RNA of the P1 helix into solution containing Pyrrolo-C labeled 5’ strand RNA of the P1 helix. The fluorescence signal is recovered by adding excessive unlabeled 5’ strand RNA of the P1 helix (for the final (blue) data point, 8 µM unlabeled 5’ strand is added as a competitor). Schematic of the assay is shown at the left. (b) Equilibrium dialysis experiment to verify SAM binding of the WT AAT RNA construct with a short P1 helix (8 base pairs). The equilibrium dialysis experiment was performed with two different SAM concentrations—the RNA is in excess with 100 nM SAM or RNA to SAM is in 1:1 ratio with 1 µM SAM. (c) Experiment to mimic the effects of SAM on shifting the conformational state towards the “OFF” form. Addition of 20 µM SAM to complex of reporter with aptamer containing partial P1 helix, results in partial recovery of fluorescence due to displacement of the reporter. (d) Control experiment showing that SAM does not displace the reporter from hybridization to a construct that cannot bind SAM due to full truncation of 5’ P1 helix-forming residues. (e) Same experiment as (c) but with an RNA construct with full wild type P1 helix. In this case no recovery of fluorescence is observed when SAM is added.