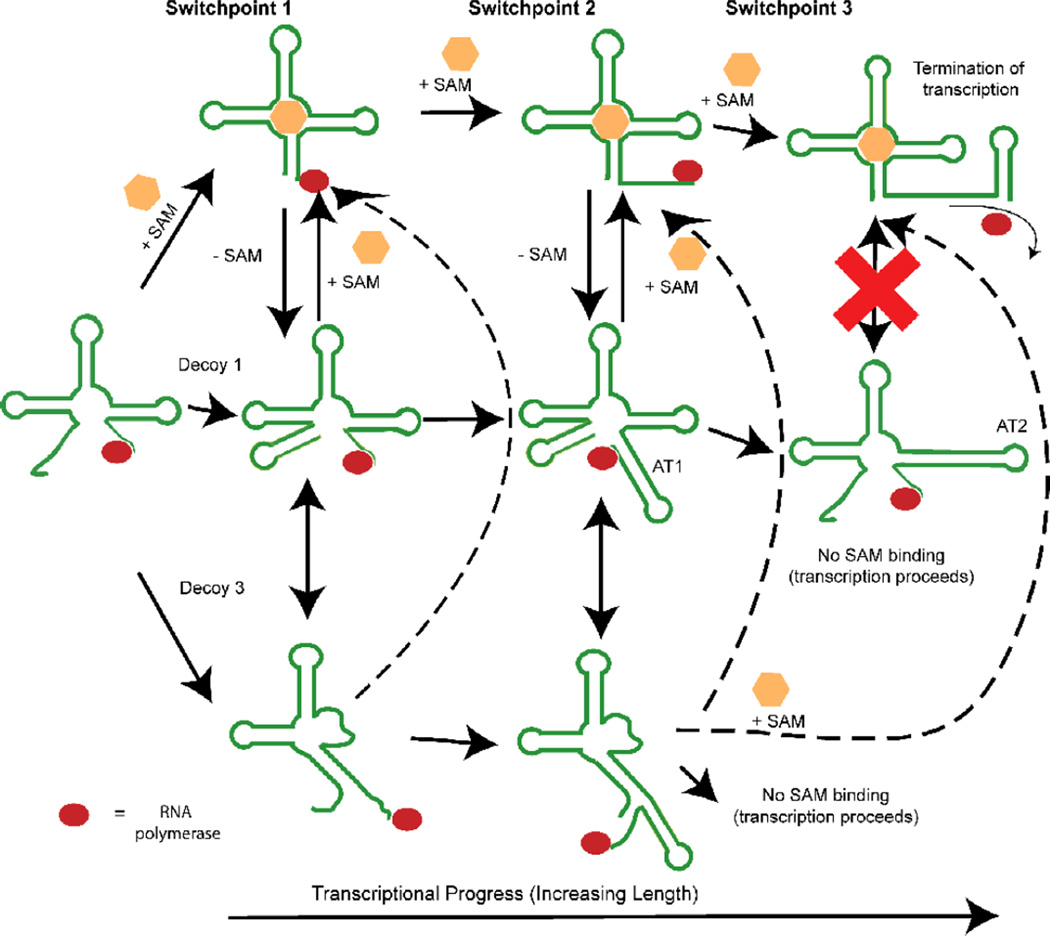
Figure 9. Schematic illustration of three conformational decision points during the synthesis of the metF SAM-I riboswitch.
The first decision point determines whether 5’ residues engage in distal interactions, leading to formation of a P1 helix, or whether they are sequestered by “decoy” purine residues in junction regions. The second decision point involves a competition between P1 and AT1 helix formation. The final decision point, which is not apparent in the yitJ SAM-I riboswitch, converts the AT1 helix to an AT2 helix. Up to this final switch leading to the formation of the AT2 helix and the transcription ON state (lower right), it is proposed that SAM binding could readily perturb the equilibrium towards the transcription OFF state (upper right). Dotted arrows link decoy 3 to SAM-bound states, because no data is available to indicate the whether decoy 3 formation is reversible upon SAM binding. RNA polymerase is represented by a red oval.