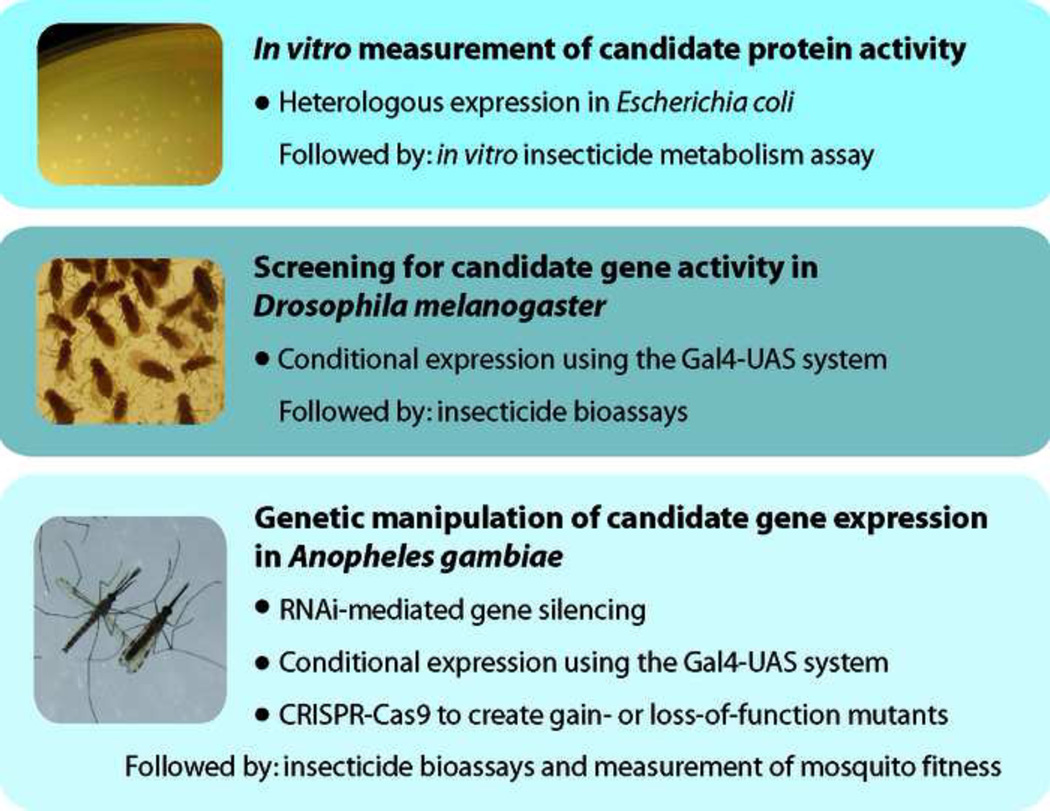
Figure 3. Strategies for functional validation of novel resistance-associated candidate genes or allelic variants.
Validation of a resistance candidate is often performed using multiple in vitro and in vivo strategies, however the choice of organism may depend upon cost, timescale, and the availability of insect culturing facilities. Two transformation systems that have the potential for high-throughput screening of candidates are Gal4-UAS and CRISPR-Cas9 [50, 51]. Gal4-UAS allows for conditional transgene expression, as your gene of interest is under the transcriptional control of Gal4 binding sites. The Gal4 yeast transactivator is encoded by a separate transgene containing user-selected regulatory sequences. Your gene of interest is expressed only when these two transgenes are joined in a single organism through genetic crosses. Alternatively, the CRISPR-Cas9 transformation system can be used if the desired outcome is a site-specific mutation or transgene insertion. When provided either in vivo or in vitro, the Cas9 nuclease creates double-stranded breaks in genomic DNA sequences complementary to a single guide RNA. These breaks are most commonly repaired through non-homologous end joining, which results in short insertions and deletions. If a plasmid DNA donor containing regions of homology is provided, homology-directed repair can result in insertion of donor sequence.