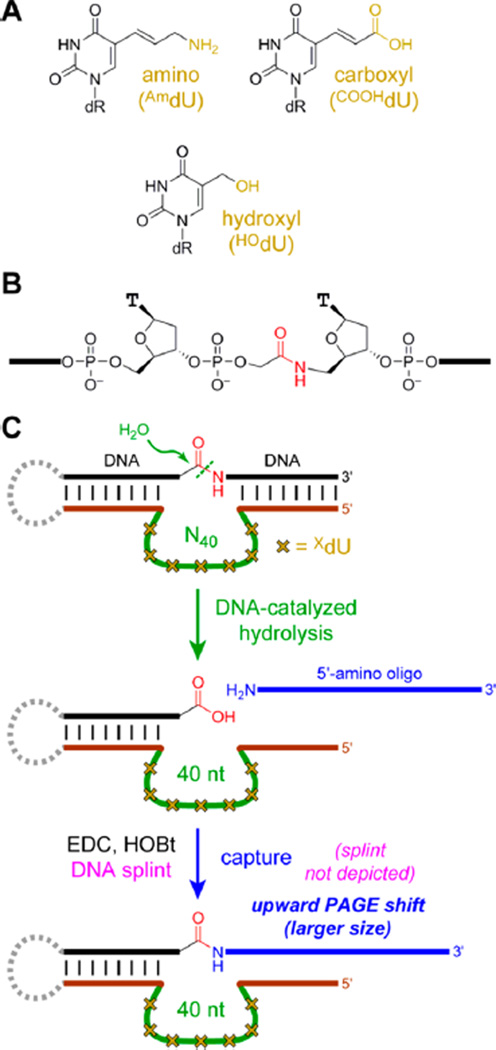
Figure 1.
In vitro selection using modified DNA to achieve amide hydrolysis. (A) Modified XdUTP nucleosides used in this study (dR = deoxyribose). (B) Amide substrate. (C) Key steps of in vitro selection, in which DNA-catalyzed amide hydrolysis is followed by DNA-splinted capture of the revealed carboxylic acid with a 5′-amino oligonucleotide. The DNA modifications in the initially random (N40) region are denoted schematically by yellow crosses. Not depicted is PCR after the capture step to form the reverse complement of the captured DNA sequences (without modified XdU), followed by primer extension (with modified XdUTP) to synthesize the DNA pool for the next round, now enriched in catalytically active sequences. See SI text and Figure S1 for selection details.