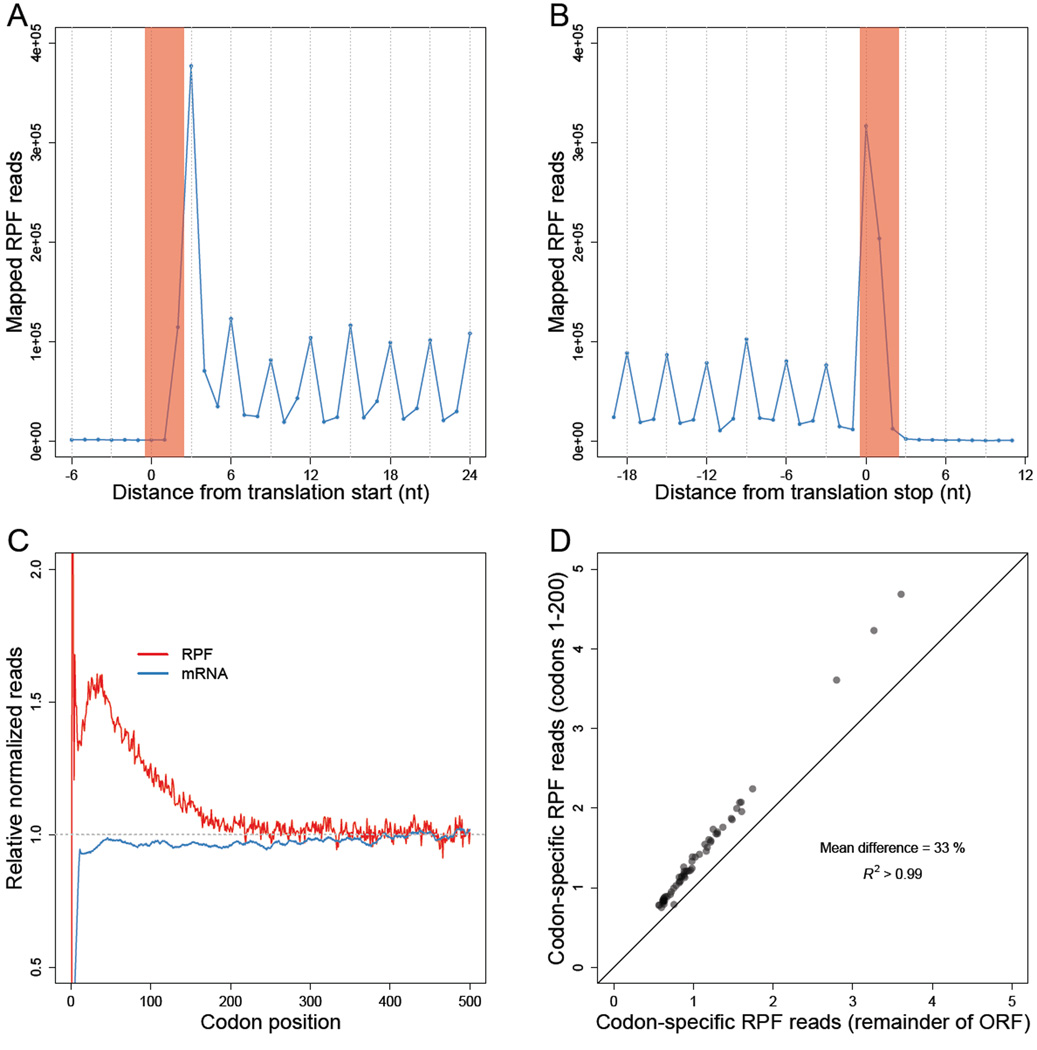
Figure 1. Less perturbed RPFs reveal a codon-independent 5' ramp.
(A–B) Metagene analyses of RPFs. Coding sequences were aligned by their start (A) or stop (B) codons (red shading). Plotted are the numbers of 28–30-nt RPF reads with the inferred ribosomal A site mapping to the indicated position along the ORF.
(C) Metagene analyses of RPFs and RNA-seq reads (mRNA). ORFs with at least 128 total mapped reads between ribosome-profiling (red) and RNA-seq (blue) samples were individually normalized by the mean reads within the ORF, and then averaged with equal weight for each codon position across all ORFs (e’j Eqn S10 and h’j Eqn S14).
(D) Comparison of codon-specific RPFs as a function of the 5' ramp. For each of the codons, densities of RPFs with ribosomal A sites mapping to that codon were calculated using either only the ramp region of each ORF (codons 1–200) or the remainder of each ORF (v5k Eqn S16 and v3k Eqn S17, respectively). The diagonal line indicates the result expected for no difference between the two regions.
See also Figure S1.