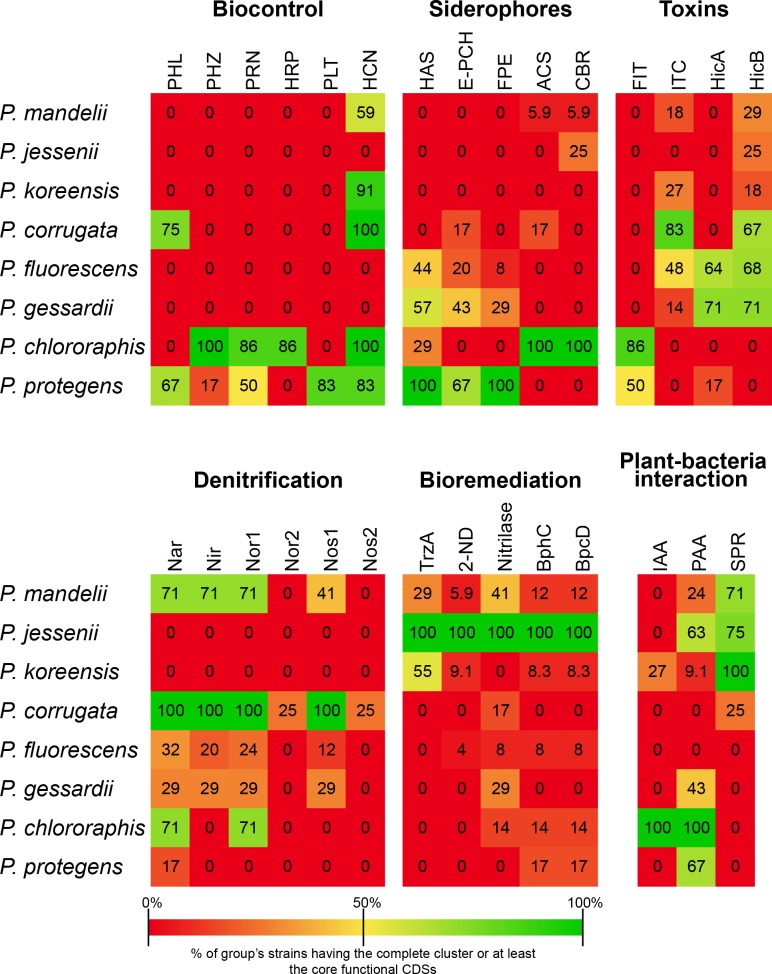
Fig 8. Distribution of the selected group-defining clusters in the groups from the P. fluorescens complex.
Numbers within the boxes represent the percentage of strains from each group having the complete gene cluster or at least the core functional CDSs. PHL: DAPG (2,4-diacetylphloroglucinol) biosynthesis (PhlABCDEFGH); PHZ: phenazine biosynthesis (PhzABCDEFGOIR); PRN: Pyrrolnitrin biosynthesis (PrnABCD); HRP: 2-hexyl, 5-propyl resorcinol biosynthesis (DarABCRS); PLT: Pyoluteorin biosynthesis (PltABCDEFGIJKLMNOPRZ); HCN: hydrogen cyanide (HcnABC); HAS: Hemophore biosynthesis (HasADEFIRS); E-PCH: Enantio-pyochelin biosynthesis (PchABCDEFHIKR); PFE: Ferric- enterobactin receptor (PfeARS); ACS: Achromobactin biosynthesis (AcsABCDEF+YhcA); CBR: Achromobactin transport (CbrABCD); Nar: Nitrate reductase (NarGHIJKLUX); Nir: Nitrite reductase (NirCDEFGHJLM1NOPS+NorQ); Nor: Nitric oxide reductase (NorBCD); Nos: Nitrous oxide reductase (NosDFLRYZ); FIT: FitD toxin (FitABCDEFGH); ITC: Insecticidal toxin complex; 2-ND: 2-nitropropane dioxigenase; IAA: Indole-3-acetic acid metabolism (IaaMH); PAA: Phenylacetic acid catabolism (PaaABCDEFGHIJKLNWXY); SPR: spermidin biosynthesis (S-adenosylmethionine decarboxylase and spermidine synthase).