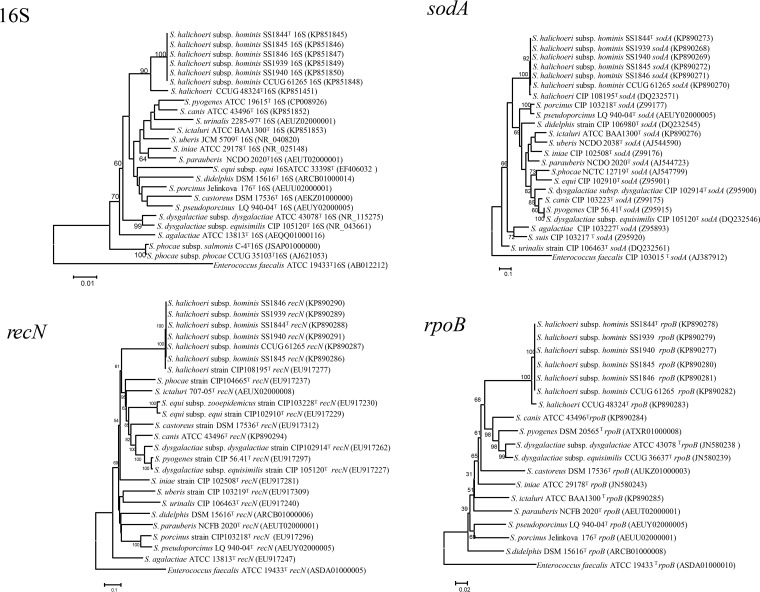
FIG 1.
Phylogenetic trees inferred from comparison of a 1,456-bp portion of the 16S rRNA gene, a 3,516-bp portion of the rpoB gene, a 417-bp portion of the sodA gene, and a 1,248-bp portion of the recN gene from Streptococcus species. The evolutionary history was inferred using the neighbor-joining method (12). Bootstrap percentage values of ≥50 from 1,000 replicates are shown at nodes (13). The evolutionary distances were computed using the maximum composite likelihood method (20) and are in units of the number of base substitutions per site. Evolutionary analyses were conducted in MEGA6 (11). No significant differences were noted in the tree topologies when maximum likelihood statistical methods were used with either Kimura 2-parameter or general-time-reversible nucleotide substitution models.