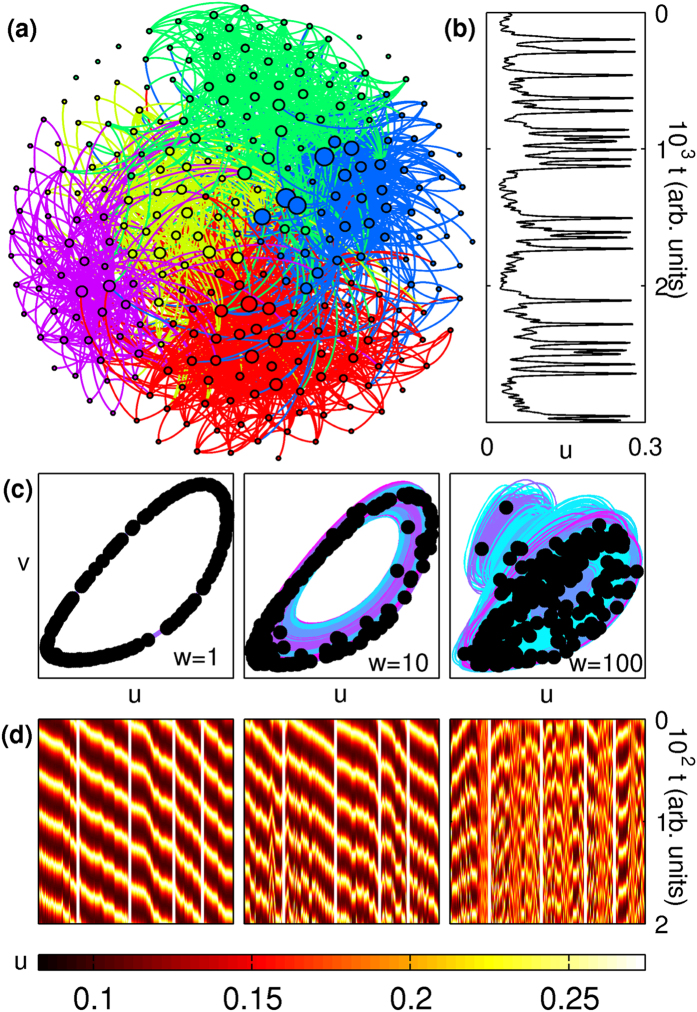
Figure 1.
(a) The directed network of connections between regions of the Macaque brain, adapted from a recent study20 (see Methods for more details). The size of each node is proportional to its total degree and the colors distinguish the modules (characterized by significantly higher intra-connection density and obtained using a partitioning algorithm39). The color of each link corresponds to that of the source node. We have used the Fruchterman-Reingold algorithm, as implemented in Gephi40, for placing the nodes of the network in a two-dimensional plane so that all links are of approximately equal length and cross each other minimally. This force-directed placement of nodes seems to clearly segregate the modules obtained with the partitioning algorithm used here. (b) Time series of the excitatory component of a typical node in this network with coupling strength w = 500, where each node is modeled as a Wilson-Cowan oscillator. (c) Phase space projections of the oscillators, obtained for different coupling strengths, where the filled circles represent the location of each oscillator at a time instant. The panels here are scaled individually for better visualization. (d) Time-series of the excitatory component u, for the corresponding values of w used in the panels directly above. The nodes i are arranged according to their modules (demarcated by white lines).