Abstract
Protein kinase D2 (PKD2) is a serine and threonine kinase that is activated in T cells by diacylglycerol and protein kinase C in response to stimulation of the T cell receptor (TCR) by antigen. We quantified the activation of PKD2 at the single-cell level and found that this kinase acts as a sensitive digital amplifier of TCR engagement, enabling CD8+ T cells to match the production of inflammatory cytokines to the quality and quantity of TCR ligands. There was a digital response pattern of PKD2 activation in response to TCR engagement, such that increasing the concentration and potency of TCR ligands increased the number of cells that exhibited activated PKD2. However, for each cell that responded to TCR stimulation, the entire cellular pool of PKD2 (~400,000 molecules) was activated. Moreover, PKD2 acted as an amplification checkpoint for antigen-stimulated digital cytokine responses and translated the differential strength of TCR signaling to determine the number of naïve CD8+ T cells that became effector cells. Together, these results provide insights into PKD family kinases and how they act digitally to amplify signaling networks controlled by the TCR.
INTRODUCTION
The mammalian serine and threonine protein kinase D (PKD) family consists of three different, but closely related, serine kinases (PKD1, PKD2, and PKD3), which integrate diacylglycerol (DAG) and protein kinase C (PKC) signaling to control diverse biological processes in multiple cell lineages. For example, PKD1 is essential for normal embryonic development (1), whereas PKD2 has an important role in adult mice to control the function of lymphoid cells during adaptive immune responses (2, 3). The activation of PKDs is initiated by the binding of polyunsaturated DAGs to N-terminal regulatory domains in the kinases, but is completed and stabilized by the DAG-dependent, PKC-mediated phosphorylation of two serine residues within the conserved PKD catalytic domain (Ser707 and Ser711 for murine PKD2) (4, 5). PKC-phosphorylated PKDs are catalytically active in the absence of continued binding of DAG, and they do not need to be localized to the plasma membrane to remain active (6). The allosteric regulation of PKDs by PKC-mediated phosphorylation thus affords a mechanism for these molecules to act as signal amplifiers that transduce signals from receptor-mediated increases in DAG and PKC from the cell membrane to the interior of the cell.
PKD2, but not PKD1, is selectively found in lymphocytes (2). PKD2 is required for signaling initiated by the T cell antigen receptor (TCR) in mature peripheral T lymphocytes (3). Stimulation of the TCR by peptide–major histocompatibility complexes (pMHCs) on the surface of antigen-presenting cells (APCs) initiates T cell proliferation (a process known as clonal expansion) and differentiation (7). Naïve T cells are highly sensitive to antigen, because only a few pMHC complexes are sufficient to stimulate the network of signaling pathways required for the differentiation of naïve T cells into effector T cells (8, 9). How TCR-mediated signaling is amplified to transduce signals that sustain T cell proliferation and control the size of the pool of effector T cells is thus a key question. Accordingly, it is important to identify the critical signaling molecules that control amplification steps in T cells because these will be relevant targets for therapeutic intervention.
In this context, the TCR is coupled through cellular tyrosine kinases to signaling responses that generate key “second messengers,” including DAG (10). A crucial role for DAG in controlling the sensitivity of TCR responses is evident in T cells that lack DAG kinases (enzymes that phosphorylate DAG to terminate its signaling), which show enhanced responsiveness to TCR stimulation (11, 12). As discussed earlier, one DAG-activated signaling molecule that is important for T cell activation is PKD2. This kinase binds to DAG with high affinity (13) and is highly abundant in peripheral T cells (2), and thus has the potential to be a sensitive sensor of TCR occupancy. Moreover, the biochemistry of PKD2 activation by PKC-mediated phosphorylation enables this kinase to transduce signals from the plasma membrane to the cytosol. Indeed, during the sustained response to TCR engagement, phosphorylated and active PKD2 molecules are localized in the cytosol (6).
In vitro studies indicate that PKD2 is important for proinflammatory cytokine production by antigen-activated T lymphocytes (2, 3). In this respect, it is increasingly recognized that the recruitment of naïve T cells into a pool of activated cells that switch on cytokine production depends on the ability of an individual T cell to sense the strength of the TCR ligand and initiate digital on and off sensitive responses that amplify TCR signaling (14, 15). Does PKD2 mediate a sensitive response to TCR ligands? To answer this question, a number of issues need to be resolved. First, does PKD2 show a digital or analog response to TCR stimulation? Second, does PKD2 activation interpret the quality or quantity of the TCR ligand? And third, does PKD2 control digital or analog responses in T cells? Here, we addressed these issues and explored the role of PKD2 as an effector of DAG and PKC in T cells in experiments with mice deficient in PKD2 (Prkd2−/−) and mice that express a variant PKD2 that cannot be phosphorylated by PKC (Prkd2S707A/S711A), and hence cannot amplify or transduce DAG- and PKC-dependent signals into the cell interior (2). Our experiments showed that PKD2 acts as an ultrasensitive digital amplifier of TCR signaling that determines the percentage of cytokine-producing cells within the T cell population and the size of the T cell effector pool. These data afford insights about PKD2 and how its role as a PKC substrate enables PKD2 to function as a signal amplifier to mediate cellular responses in CD8+ T cells.
RESULTS
Antigen-stimulated T cells exhibit digital activation of PKD2
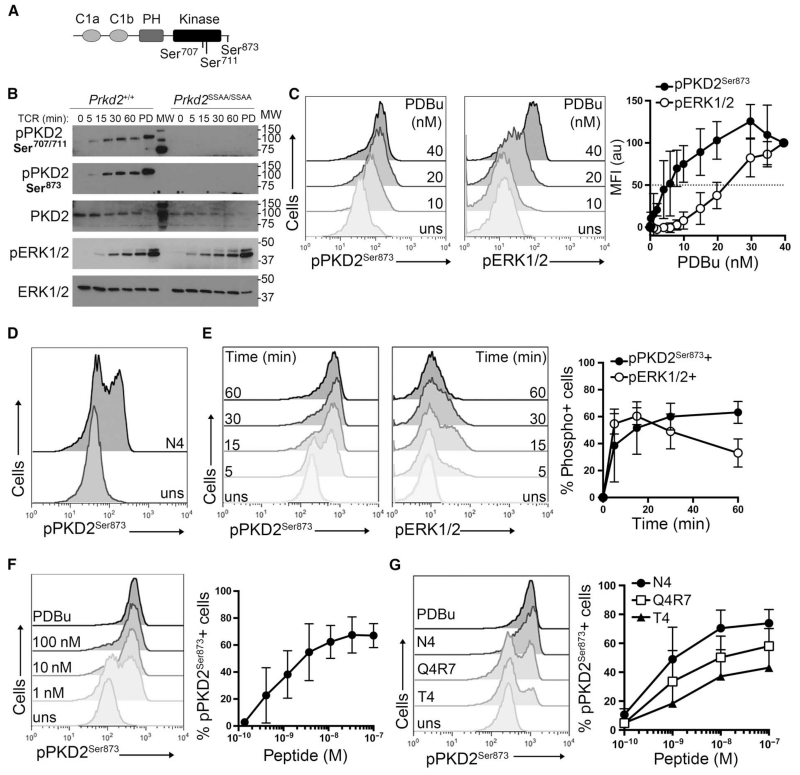
PKD2 activation is initiated by the binding of DAG to an N-terminal regulatory domain in the kinase, but activation is completed by the PKC-mediated phosphorylation of Ser707 and Ser711 within the activation loop of the catalytic domain of PKD2 (Fig. 1A). To assess the importance of these phosphorylation events for PKD2 activity, we monitored the activity of PKD2 by Western blotting analysis with antisera against PKD2 phosphorylated at Ser873 (pPKD2Ser873) (Fig. 1B), a known PKD2 autophosphorylation site (Fig. 1A). In these experiments, we used naïve OT-I TCR transgenic T cells, a genetically identical monoclonal population of CD8+ T cells that express a TCR that recognizes the chicken ovalbumin (OVA) peptide, SIINFEKL (N4), presented by the type I MHC molecule H-2Kb on APCs. These experiments showed that in naïve T cells, stimulation of the OT-I TCR with its cognate ligand SIINFEKL (N4) induced a rapid and sustained activation of PKD2 (Fig. 1B). The ability of the OT-I TCR to activate PKD2 was markedly reduced in T cells expressing PKD2 S707A/S711A (Prkd2SSAA/SSAA), which cannot be phosphorylated by PKCs (2, 3). As a positive control for TCR signaling, we found that the TCR-stimulated activation of extracellular signal–regulated kinase 1/2 (ERK1/2) was not altered in OT-I Prkd2SSAA/SSAA CD8+ T cells (Fig. 1B).
Fig. 1. PKD2 displays a digital pattern of activation in response to TCR stimulation.
(A) Schematic representation of PKD2 structure. C1a and C1b are DAG-binding domains; PH, pleckstrin homology domain; Kinase, catalytic domain; Ser704 and Ser711, T loop serine residues; Ser873, autophosphorylation serine residue. (B) Naïve OT-I CD8+ cells from Prkd2+/+ and Prkd2SSAA/SSAA mice were stimulated through the TCR with 100 nM N4 peptide for the indicated times or were treated with PDBu (PD) for 1 hour. Cell lysates were then analyzed by Western blotting with antibodies against the indicated proteins. TCR-stimulated ERK1/2 activation was used as a positive control. Western blots are representative of three experiments. (C to G) Intracellular flow cytometric analysis of the phosphorylation of PKD2-Ser873 (pPKD2Ser873) and ERK1/2 (pERK1/2) in naïve OT-I CD8+ T cells. (C) Cells were left unstimulated (uns) or were stimulated with the indicated amounts of PDBu for 15 min. Representative histograms show PKD2 and ERK1/2 phosphorylation. Graph shows the mean fluoresce intensity (MFI) ± SD for pPKD2Ser873 and pERK1/2 expressed in arbitrary units (au) relative to the MFI obtained for the maximal dose. Data are from three independent experiments. (D) Cells were left unstimulated or were stimulated with 0.3 nM N4 peptide for 1 hour and then were analyzed for PKD2Ser873 phosphorylation. Histograms are representative of 10 independent experiments. (E) Cells were left unstimulated or were stimulated with 10 nM N4 peptide for the indicated times. Representative histograms show PKD2 and ERK1/2 phosphorylation. Graph shows the mean ± SD of the percentages of cells positive for pPKD2Ser873- and pERK1/2-positive cells. Data are from four independent experiments. (F) Cells were left unstimulated or were treated with the indicated concentrations of N4 peptide or with PDBu (20 ng/ml) for 30 min. Representative histograms show pPKD2Ser873 staining. Graph shows the mean percentage ± SD of cells positive for pPKD2Ser873. Data are from five independent experiments. (G) Cells were treated with increasing concentrations of the N4, Q4R7, and T4 peptides for 1 hour. Representative histograms show pPKD2Ser873 staining in cells treated with 10 nM peptide. Graph shows the mean percentage + SD of cells positive for pPKD2Ser873 in response to the indicated peptide concentrations. Data are from three independent experiments.
Phorbol esters, such as phorbol 12,13-dibutyrate (PDBu), which act as pharmacological mimetics of DAG, stimulate a stoichiometric phosphorylation and activation of the total cellular pool of PKDs. In this respect, PKD2 activity in the TCR-stimulated T cells was lower than that seen in the PDBu-treated T cells (Fig. 1B). This could reflect that TCR stimulation is unable to activate the total pool of PKD2 found within each individual T cell. Alternatively, the lower amount of active PKD2 seen at the population-level analysis of a Western blot could reflect the possibility that not all T cells within the naïve T cell pool could activate PKD2 in response to TCR stimulation. To distinguish between these possibilities, we used a single-cell, flow cytometry–based protocol to quantify PKD2 activity. The antiserum against PKD2 phosphorylated at Ser873 (pPKD2Ser873) accurately monitors PKD2 activity in flow cytometry–based assays and enables quantification of PKD2 activity at the single-cell level (3). The sensitivity of this assay was assessed in a PDBu dose-response curve for PKD2 activation (Fig. 1C). In these experiments, we also used flow cytometry to monitor the PDBu dose response for ERK1/2 activation because the analysis of ERK1/2 phosphorylation by flow cytometry has been used extensively to monitor DAG-mediated signaling pathways in T cells (16, 17). These experiments showed that PKD2 activation was sensitive to PDBu, with a 50% maximal response achieved with 10 nM PDBu, compared to the 25 nM PDBu that was required to achieve a similar response for ERK1/2 activation (Fig. 1C). These experiments also showed that increasing the concentration of PDBu promoted a gradual shift in pPKD2Ser873 staining (Fig. 1C). These data suggest that PDBu activates PKD2 in the total pool of naïve T cells in an analog manner.
We then examined the characteristics of TCR-stimulated activation of PKD2. The key question was whether the TCR stimulated an analog or digital PKD2 activation response in naïve OT-I CD8+ T cells. The data showed that after stimulation of the OT-I TCR with its cognate ligand SIINFEKL (N4), two distinct cell populations were detected with respect to pPKD2Ser873 staining: one population had active PKD2 or was “on,” whereas the other population had no detectable PKD2 activity or was “off” (Fig. 1D). This on-off state of active PKD2 suggested that there was a digital response to TCR stimulation, which was in contrast to the graded response in PKD2 activation that was observed for PDBu (Fig. 1, C and D). We next examined the kinetics of PKD2 activation after TCR stimulation in parallel with that of ERK1/2 activation, because ERK1/2 exhibits a digital response pattern in naïve T cells (16). In these experiments, we detected on and off activation states for both PKD2 and ERK1/2, which are characteristic of digital behavior (Fig. 1E). These experiments also confirmed previous results showing that the TCR-dependent activation of ERK1/2 is transient (16). In comparison, the TCR-dependent activation of PKD2 was rapid and sustained (Fig. 1E), and the effect of prolonging the time of TCR stimulation was to increase the percentage of cells that had activated PKD2. We also noted that the intensity of the pPKD2Ser873 staining among the population of cells with active PKD2 did not change, indicating that similar numbers of PKD2 molecules were activated in the responding population at the different time points.
A characteristic feature of a digital (or bimodal) response is that increasing the concentration of the stimulus increases the percentage of responding cells, which is in contrast to a graded or analog response in which increasing the concentration of the stimulus gradually increases the response in individual cells. Therefore, we examined in detail the digital behavior of PKD2 activation in naïve OT-I T cells as they responded to increasing concentrations of TCR ligand (pMHC). These experiments showed that the percentage of CD8+ T cells that contained active PKD2 was determined by the concentration of SIINFEKL (N4) peptide used, but that the amount of active PKD2 among the responding cells remained constant regardless of the increased pMHC concentration (Fig. 1F). We did not detect cells with intermediate amounts of pPKD2Ser873 in response to TCR stimulation. In these experiments, we used cells stimulated with PDBu as a positive control because this pharmacological stimulus of PKD2 maximally activates the total cellular pool of PKD2. A comparison of the amounts of pPKD2Ser873 in cells stimulated either through the TCR or with PDBu showed that, independently of the peptide concentration used, the amount of active PKD2 per cell was comparable to that observed in PDBu-activated cells (Fig. 1F). These results confirm the digital behavior of PKD2 activation in response to TCR stimulation, and showed that in individual T cell responders, the total cellular pool of PKD2 molecules was active.
TCR ligand affinity controls the number of naïve T cells that activate PKD2
We also investigated how the pMHC affinity for the TCR affected PKD2 activation in the OT-I TCR transgenic model. The peptide SIINFEKL (N4) presented by H-2Kb is a strong agonist for the OT-I TCR complex. We made use of known variants of the N4 peptide with a wide range of potencies for stimulating OT-I T cells, which have been extensively used to study how pMHC affinity for the TCR influences selection processes in the thymus (18) and activation thresholds in naïve T cells (19, 20). We selected two peptide variants derived from the original high-affinity OT-I ligand N4: SIIQFERL (Q4R7) and SIITFEKL (T4), which have intermediate and low affinity (Kd = 48 ± 9.5 nM and 55 ± 10.1 nM, respectively) for the TCR compared to that of the high-affinity N4 peptide (Kd = 3.7 ± 0.7 nM) (18). These experiments showed that the affinity of the pMHC for the TCR determined the percentage of T cells that were able to activate PKD2 (Fig. 1G). The T4 peptide, which has low affinity for the OT-I TCR, stimulated only a few T cells to activate PKD2; however, within all of the responding cells, the total pool of PKD2 molecules was activated. The percentage of T cells with active PKD2 was higher when a peptide with intermediate affinity for the TCR (Q4R7) was used, although the percentage of responding cells was still lower than that stimulated by the strong agonist N4 peptide. These data thus show that PKD2 displays a digital pattern of activation in response to TCR stimulation, and that both TCR ligand concentration and potency dictate the percentage of responding cells (Fig. 1, F and G). In individual responders, 100% of PKD2 molecules were activated in response to TCR stimulation.
PKD2 is a sensitive sensor of TCR signaling
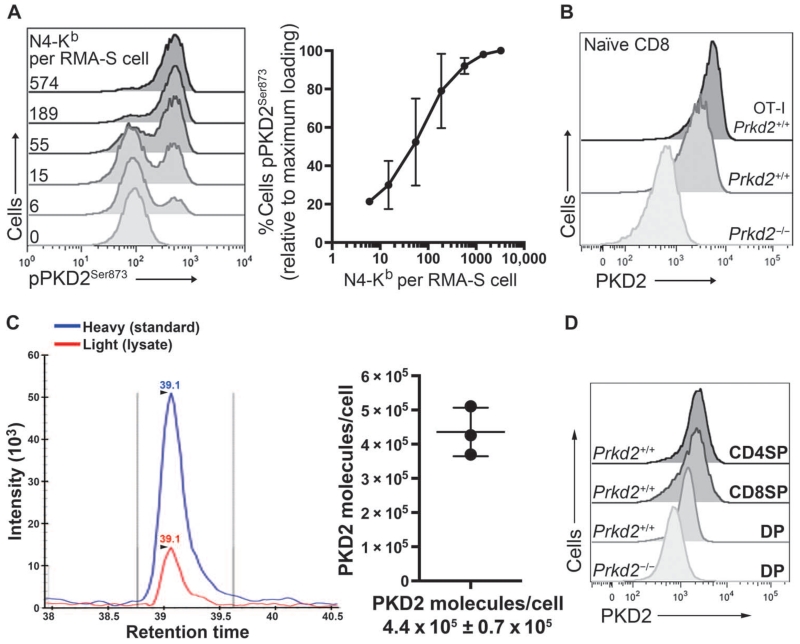
T cells are very sensitive to TCR stimulation, and very few pMHC complexes are required to initiate T cell activation processes (8, 9, 14, 16). The phorbol ester dose response for PKD2 activation (Fig. 1C) indicated that PKD2 activation could be induced in response to very low amounts of stimuli. We therefore wished to examine how many pMHC molecules were required to activate the total pool of PKD2 molecules in a single T cell. To control the amount of cognate peptide presented to T cells, we used the transporter associated with antigen processing (TAP)–deficient lymphoma RMA-S as peptide-loaded APCs. RMA-S cells efficiently present exogenously added peptide through H-2Kb MHC molecules. Moreover, an anti–N4-Kb antibody that recognizes the N4 peptide bound to H-2Kb can be used to quantify the number of pMHC molecules on the cell surface of the APC (16). We loaded RMA-S cells with different amounts of N4 peptide and determined the amount of pMHC at the cell surface by comparing the MFI of phycoerythrin (PE)–conjugated anti–N4-Kb antibody staining to that of a known PE standard curve (fig. S1). RMA-S cells loaded with different amounts of N4 peptide were then mixed with naïve OT-I CD8+ T cells, and PKD2 activation was assessed by flow cytometry. Peptide loading of the RMA-S cells showed a normal Gaussian distribution, and loading an average of 6 ± 103 pMHC complexes per RMA-S cell was sufficient to activate the total pool of PKD2 in an individual T cell (Fig. 2A and fig. S1). Moreover, increasing the density of pMHC ligands on the APC increased the percentage of T cells within the population that exhibited activated PKD2 (Fig. 2A). These data suggest that PKD2 activation is a sensitive digital sensor of TCR signaling in response to peptide-loaded APCs.
Fig. 2. PKD2 is a sensitive amplifier of TCR signaling.
(A) Naïve OT-I CD8+ T cells were stimulated for 2 hours with RMA-S loaded with the indicated amounts of N4 peptide and then assessed by flow cytometry. Representative histograms show pPKD2Ser873 staining. Inset: numbers indicate the number of pMHC complexes (N4-Kb) presented on the surface of each RMA-S cell. Graph shows the mean percentage ± SD of cells that were positive for pPKD2Ser873 relative to the percentage of cells that occurred with the maximal dose of pMHC. Data are from three independent experiments. (B) Flow cytometric analysis of PKD2 abundance in naïve OT-I CD8+ T cells and polyclonal CD8+ T cells with anti-PKD2 antisera. Staining of Prkd2−/− cells is shown as a negative control for antisera specificity. Histograms are representative of two independent experiments. (C) Absolute quantification of PKD2 in naïve OT-I CD8+ T cells as assessed by SRM. Histogram shows the peak intensities of co-eluted light (endogenous) and heavy (standard) PKD2654–664 peptides. Peak intensities of six transitions were used to back-calculate the number of PKD2 molecules per cell. Graph indicates the number of PKD2 molecules per cell expressed as an average ± SD. Data are from three biological replicates. (D) Flow cytometric analysis of PKD2 abundance in cells at different stages of development in the thymus in polyclonal mice. Histograms are representative of two independent experiments.
Amplification of TCR signals by PKD2 activation
To quantify the extent of signal amplification associated with the activation of the total cellular pool of PKD2 by small amounts of pMHC complexes, it was necessary to determine the number of PKD2 molecules present in peripheral T cells. Flow cytometric analysis of PKD2 abundance with a pan-PKD antiserum revealed a Gaussian distribution of PKD2 molecules within the monoclonal, naïve OT-I CD8+ T cell population (Fig. 2B). We then used a targeted mass spectrometry (MS) technique known as selected reaction monitoring (SRM) to quantify PKD2 abundance in naïve OT-I CD8+ T cells, because this method has the potential to yield absolute quantification of target peptides from a particular protein in a complex sample, such as a peptide mixture derived from a trypsin digestion of total cell lysates. SRM also has the potential to detect, in principle, proteins within the low-attomole range of abundance (21), and absolute proteomic quantitation is achieved by spiking the experimental sample with known concentrations of synthetic, heavy isotopologues of target peptides and then performing liquid chromatography (LC)–MS/MS. Thus, by SRM, heavy and light versions of the target peptides are analyzed by MS simultaneously, and the abundance of the target peptide in the experimental sample is compared to that of the heavy peptide and back-calculated to the initial concentration of the heavy standard. PKD2654–664 peptide (FLITQILVALR) was selected as optimal for PKD2 quantification by SRM (see Materials and Methods). A known concentration of synthetic PKD2654–664 peptide labeled with heavy isotopes was mixed with the trypsin digest obtained from naïve OT-I CD8+ T cells and analyzed by SRM. These experiments indicated that naïve OT-I CD8+ T cells had, on average, 370,000 to 500,000 molecules of PKD2 per cell (Fig. 2C). Accordingly, the activation of PKD2 is a markedly sensitive means of signal amplification for the TCR given that up to 500,000 molecules of PKD2 are activated by stimulation with small amounts of pMHC per T cell (Fig. 2A).
A comparison of PKD2 abundance in naïve OT-I CD8+ T cells and that in naïve polyclonal CD8+ T cells revealed that the abundance of PKD2 in the OT-I TCR transgenic T cells was almost twofold greater than that in polyclonal CD8+ T cells (Fig. 2B). This difference led us to consider that the abundance of PKD2 in naïve T cells might be set while the T cells undergo positive selection in the thymus. Our flow cytometric analysis of PKD2 abundance in thymic subsets (Fig. 2D) is consistent with this hypothesis, because we observed that there was a substantial increase in PKD2 abundance in T cells as they differentiated from the less mature CD4+CD8+ double-positive (DP) cell population into the mature CD4+ and CD8+ single-positive (SP) cells.
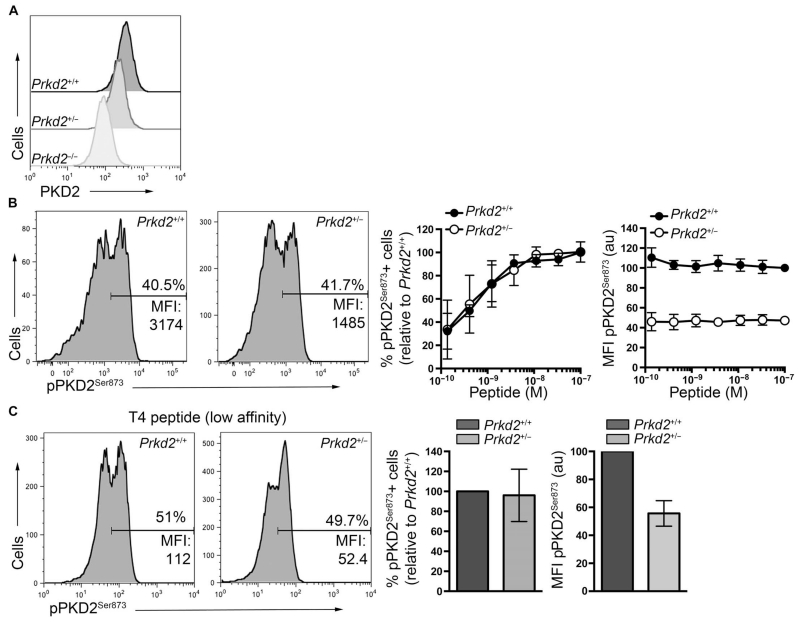
Why do T cells have such a high quantity of PKD2? One possibility is that the abundance of PKD2 enables the T cell to sense small changes in the amount of cellular DAG. To explore this possibility, we compared the PKD2 activation responses of OT-I Prkd2+/+ T cells to those of OT-I Prkd2+/− T cells, which have twofold less PKD2 abundance (Fig. 3A). We analyzed the percentages of OT-I Prkd2+/+ and OT-I Prkd2+/− T cells that exhibited activated PKD2 as they responded to increasing amounts of N4 peptide (Fig. 3B). These data showed that a twofold reduction in the cellular abundance of PKD2 did not change the sensitivity of T cells in terms of their ability to activate PKD2 in response to small amounts of pMHC complexes (Fig. 3B). Thus, the percentages of OT-I Prkd2+/+ and OT-I Prkd2+/− T cells that activated PKD2 in response to TCR stimulation were identical (Fig. 3B, middle), although the responding OT-I Prkd2+/− T cells had a twofold decrease in the amount of active PKD2 compared to that in OT-I Prkd2+/+ cells (Fig. 3B, right). We also noted that decreasing the cellular concentration of PKD2 did not influence the ability of T cells to discriminate between TCR ligands with different affinity. The percentage of T cells that activated PKD2 in response to low- rather than high-affinity pMHC was thus identical for OT-I Prkd2+/+ and OT-I Prkd2+/− T cells; however, as before, the amount of active PKD2 in the Prkd2+/− cells was twofold lower than that in Prkd2+/+ cells (Fig. 3C).
Fig. 3. Effect of PKD2 abundance on the activation of naïve OT-I CD8+ T cells.
(A) Flow cytometric analysis of PKD2 abundance in naïve OT-I CD8+ T cells from Prkd2+/+, Prkd2+/−, and Prkd2−/− mice. Histograms are representative of two independent experiments. (B) Naïve OT-I CD8+ T cells from Prkd2+/+ and Prkd2+/− mice were stimulated with increasing amounts of N4 peptide for 1 hour and then were analyzed by flow cytometry. Left: Representative histograms show pPKD2Ser873 staining in the indicated cells after stimulation with 1 nM N4 peptide. Inset numbers indicate the percentages of cells that were positive for pPKD2Ser873, as well as the MFI for pPKD2Ser873 in the positive cells. Graphs show the percentages of cells positive for pPKD2Ser873 (left graph) and the MFI of pPKD2Ser873 in positive cells (right graph) relative to Prkd2+/+ cells treated with the maximal concentration of N4 peptide. Data are means ± SD from three independent experiments. (C) Naïve OT-I CD8+ T cells from Prkd2+/+ and Prkd2+/− mice were treated with 100 nM T4 peptide for 1 hour and then were analyzed by flow cytometry. Representative histograms show pPKD2Ser873 staining in the indicated cells. Graphs show the percentages of cells positive for pPKD2Ser873 (left graph) and the MFI of pPKD2Ser873 in positive cells (right graph) relative to Prkd2+/+ cells. Data are means ± SD from four independent experiments.
The amount of PKD2 matters
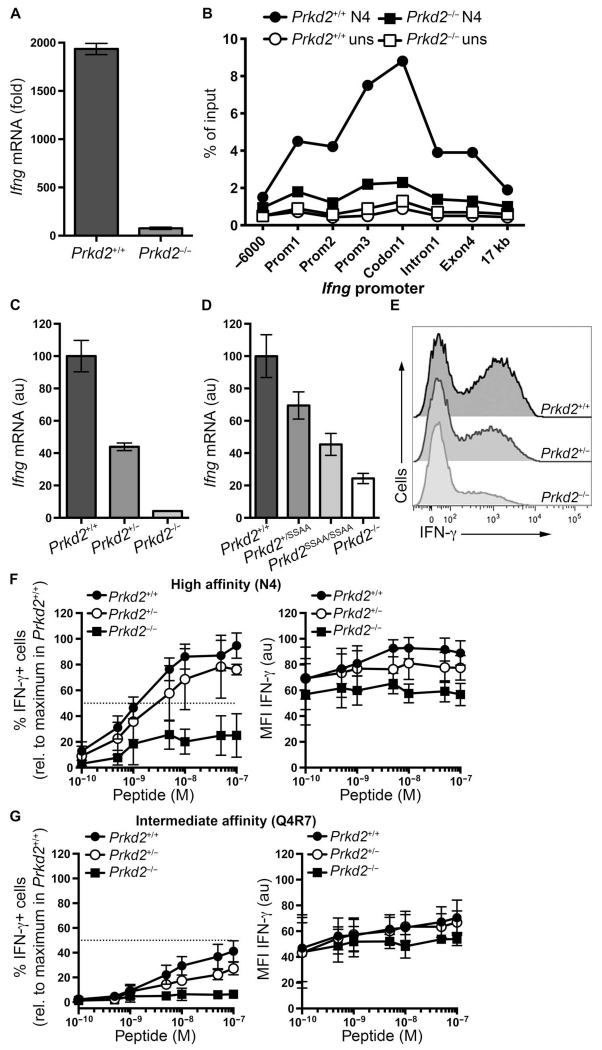
We previously showed that PKD2 is important for the TCR-stimulated production of key proinflammatory cytokines (2, 3). Consistent with these findings, we found that naïve OT-I Prkd2−/− T cells had a substantial reduction in Ifng mRNA abundance after TCR stimulation compared to that in OT-I Prkd2+/+ T cells. PKD2 was required for the expression of Ifng, because loss of PKD2 reduced the amount of RNA polymerase II (Pol II) that was recruited to the Ifng, transcription start site, and distal exons in TCR-stimulated T cells (Fig. 4B).
Fig. 4. Effect of PKD2 abundance on IFN-γ production.
(A) Naïve OT-I CD8+ T cells from Prkd2+/+ and Prkd2−/− mice treated with 2 nM N4 peptide for 5 hours were subjected to real-time polymerase chain reaction (PCR) analysis to determine the abundance of Ifng mRNA relative to that in untreated cells. Data are means ± SD of triplicate samples from one experiment and are representative of five independent experiments. (B) Naïve OT-I CD8+ T cells from Prkd2+/+ and Prkd2−/− mice treated with 10 nM N4 peptide for 6 hours were subjected to chromatin immunoprecipitation (ChIP) assays to determine the extent of Pol II binding to the Ifng promoter. Binding is presented as a percentage of input. Data are representative of two independent experiments. (C) Naïve OT-I CD8+ T cells from Prkd2+/+, Prkd2+/−, and Prkd−/− mice treated with 2 nM N4 peptide for 4 hours were subjected to real-time PCR analysis to determine the abundance of Ifng mRNA relative to that in OT-I Prkd2+/+ cells. Data are means ± SD of triplicate samples from one experiment and are representative of three independent experiments. (D) Naïve OT-I CD8+ T cells from Prkd2+/+, Prkd2+/SSAA, Prkd2SSAA/SSAA, and Prkd2−/− mice treated with 2 nM N4 peptide for 4 hours were subjected to real-time PCR analysis to determine the abundance of Ifng mRNA relative to that in OT-I Prkd2+/+ cells. Data are means ± SD of triplicate samples from one experiment and are representative of two independent experiments. (E) Naïve OT-I CD8+ T cells from Prkd2+/+, Prkd2+/−, and Prkd2−/− mice stimulated with 10 nM N4 peptide for 6 hours were subjected to intracellular flow cytometry to determine the abundance of IFN-γ. Histograms are representative of three independent experiments. (F and G) Naïve OT-I CD8+ T cells from Prkd2+/+, Prkd2+/−, and Prkd2−/− mice stimulated for 6 hours with the indicated concentrations of (F) the high-affinity peptide N4 or (G) the intermediate-affinity peptide Q4R7 were analyzed by flow cytometry to determine IFN-γ production. Left: Mean percentages ± SD of cells that were positive for IFN-γ relative to Prkd2+/+ cells treated with the maximum concentration of N4 peptide. Data are from three independent experiments. Right: Means ± SD of the MFI of IFN-γ in positive cells relative to the MFI of IFN-γ in Prkd2+/+ cells treated with the maximum concentration of N4 peptide. Data are from three independent experiments.
Is the amount of PKD2 important for TCR-dependent control of Ifng expression? We addressed this question by measuring the abundance of Ifng mRNA in TCR-stimulated OT-I Prkd2+/+, OT-I Prkd2+/−, and OT-I Prkd2−/− T cells. These experiments showed that OT-I Prkd2+/− T cells, which have half the amount of PKD2 of OT-I Prkd2+/+ cells, had only about 50% of the amount of Ifng mRNA in response to TCR stimulation (Fig. 4C). We also examined TCR-stimulated Ifng mRNA abundance in OT-I Prkd2SSAA/SSAA and OT-I Prkd2+/SSAA T cells. The loss of the PKC phosphorylation sites in PKD2 reduces its catalytic activity to about 10% of that of wild-type PKD2 (2). Our analysis of Ifng mRNA production in OT-I Prkd2SSAA/SSAA and OT-I Prkd2+/SSAA T cells, which have PKD2 variants with about 10 and 60%, respectively, of the activity of wild-type PKD2, showed that they had a corresponding reduction in their abundance of Ifng mRNA (Fig. 4D). Collectively, these data suggest that the ability of naïve CD8+ T cells to produce Ifng in response to TCR engagement is directly dependent on the quantity of active PKD2 in the T cells.
PKD2 controls the recruitment of naïve CD8+ T cells into the effector pool
Cytokine secretion by T cells is a digital response (14, 20), whereby increasing the concentration of a stimulus increases the percentage of cytokine-producing cells, but the rate of cytokine production per cell does not change with respect to stimulus concentration. We found that the number of active PKD2 molecules in TCR-stimulated T cells correlated with their Ifng mRNA abundance (Fig. 4, C and D). However, it has not been determined whether PKD2 controls the number of T cells that produce interferon-γ (IFN-γ) in response to TCR engagement or controls the rate of IFN-γ production per cell. To distinguish between these two possibilities, we used flow cytometry to assess the percentages of OT-I Prkd2+/+, OT-I Prkd2+/−, and OT-I Prkd2−/− naïve T cells that produced IFN-γ in response to activation with the high-affinity N4 peptide. These results demonstrate that reducing the amount of PKD2 by twofold caused a decrease in the number of TCR-stimulated T cells that produced IFN-γ, and that very few TCR-stimulated PKD2 null cells produced IFN-γ (Fig. 4E).
The number of naïve CD8+ T cells that produce IFN-γ in response to TCR stimulation is determined by pMHC density and potency (20). Having demonstrated that the number of active PKD2 molecules per cell correlated with the magnitude of IFN-γ production by naïve OT-I T cells (Fig. 4, C to E), we next asked whether the amount of cellular PKD2 affected T cell sensitivity to pMHC density in terms of the ability of the T cells to produce IFN-γ. To explore this possibility, we used flow cytometry to determine the percentages of naïve OT-I Prkd2+/+, Prkd2+/−, and Prkd2−/− T cells that produced IFN-γ in response to increasing concentrations of N4 peptide. The results obtained for OT-I Prkd2+/+ were consistent with previously published data (20), and showed that increasing the amount of N4 peptide increased the percentage of IFN-γ–producing cells (Fig. 4F, left) and had a relatively small effect on the amount of IFN-γ produced by the responding cells (Fig. 4F, right). These data suggest that naïve T cells face an all-or-none decision in the context of IFN-γ production, and that success at this activation checkpoint is determined by the strength of TCR engagement. The results obtained for OT-I Prkd2+/− and OT-I Prkd2−/− T cells demonstrated that the amount of cellular PKD2 positively correlated with the percentage of IFN-γ–producing cells (Fig. 4F, left), suggesting that PKD2 facilitates the progression of T cells beyond the particular signaling threshold required for IFN-γ secretion. It was also evident that the quantity of cytokine produced by the responding population was influenced by the amount of PKD2 in the cell (Fig. 4F, right).
We also examined IFN-γ production by naïve OT-I Prkd2+/+, Prkd2+/−, and Prkd2−/− T cells in response to increasing concentrations of a peptide variant (Q4R7) with intermediate affinity for the OT-I TCR (Fig. 4G). These experiments confirmed that both the density and the potency of pMHC determined the percentage of OT-I Prkd2+/+ cells that produced IFN-γ (Fig. 4, F and G, left panels). Furthermore, these data suggest that although increasing the concentration or affinity of the peptide increased the percentages of OT-I Prkd2+/− and OT-I Prkd2−/− cells that produced IFN-γ, the final outcome was determined by the amount of PKD2 present in the cells, even under conditions of TCR saturation (Fig. 4, F and G). In contrast to the results obtained from experiments with a high-affinity peptide (Fig. 4F, right), increasing amounts of an intermediate-affinity peptide barely influenced the quantity of cytokine produced by responder OT-I Prkd2+/+ cells (Fig. 4G, right). In addition, the rate of IFN-γ production in response to an intermediate-affinity peptide was not affected by the amount of cellular PKD2 (Fig. 4G, right), although the percentage of cells that produced cytokine was dependent on PKD2 (Fig. 4G, left). These experiments also showed that PKD2 was particularly critical for IFN-γ production during low-affinity TCR-pMHC interactions because only increased concentrations of a high-affinity peptide were capable of stimulating a decreased IFN-γ response in OT-I Prkd2−/− T cells (Fig. 4, F and G, left panels).
PKD2 acts as a PKC effector in CD8+ T cells in vivo
Our in vitro studies suggest that PKD2 determines the percentage of naïve CD8+ T cells that become IFN-γ producers. In this context, we previously showed that PKD2 is not required for the TCR-stimulated induction of immediate-early gene expression, but that it has a selective role in the TCR transcriptional program, and, in particular, that PKD2 couples the TCR to the induction of genes encoding multiple cytokines and chemokines (3). These data led us to predict that PKD2 would be important for T cell effector function in vivo, but this has not been assessed directly. Moreover, it has not been assessed whether the phosphorylation of PKD2 by PKC is important for T cell effector function in vivo.
To address these issues, we assessed the effector function of Prkd2SSAA/SSAA CD8+ T cells in an autoimmune diabetes model, the rat insulin promoter–membrane-bound OVA (RIP-mOVA) transgenic mice. RIP-mOVA mice express membrane-bound OVA under the control of the rat insulin promoter in β cells of pancreatic islets. Adoptive transfer of naïve OT-I CD8+ T cells into RIP-mOVA mice followed by immunization with OT-I ligands results in the rapid destruction of β cells by antigen-specific OT-I effector T cells with the subsequent development of diabetes (19, 22). Accordingly, we assessed the importance of PKD2 for CD8+ T cell effector function in OT-I Prkd2+/+ and OT-I Prkd2SSAA/SSAA CD8+ T cells by adoptively transferring them into RIP-mOVA hosts before analyzing the onset of diabetes after immunization of the mice with lipopolysaccharide (LPS) and the N4 peptide. Mice that received OT-I Prkd2SSAA/SSAA cells exhibited a statistically significant delay in the onset of diabetes compared to that in RIP-mOVA mice that received OT-I Prkd2+/+ cells, with some mice being resistant to diabetes onset (Fig. 5A).
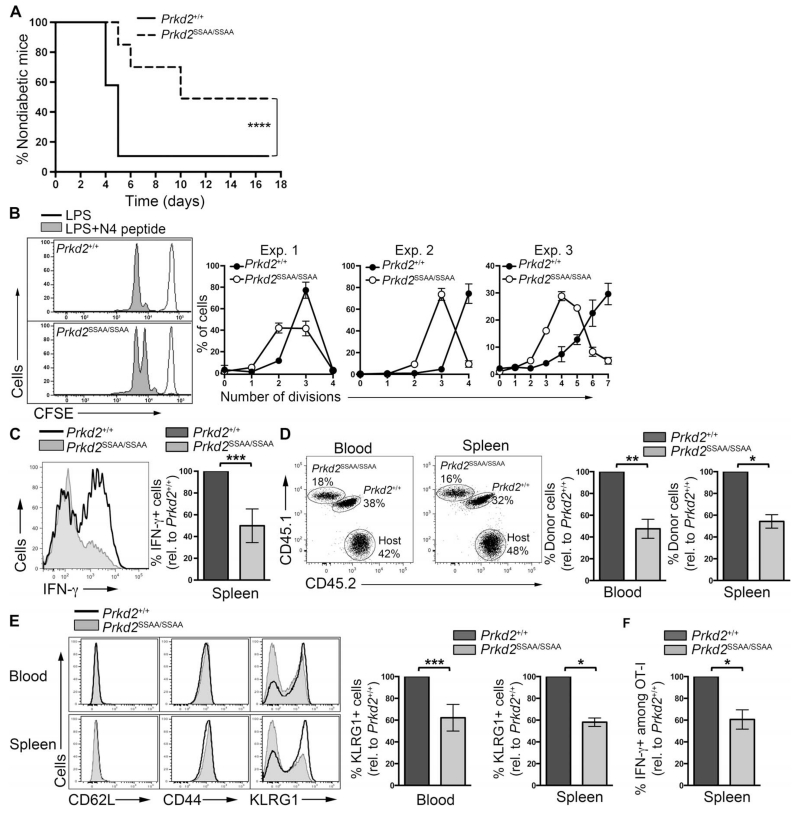
Fig. 5. PKD2 mediates CD8+ T cell–dependent immune responses in vivo.
(A) Diabetes onset in the RIP-mOVA model of autoimmune diabetes. Naïve OT-ICD8+ T cells from Prkd2+/+ and Prkd2SSAA/SSAA mice were adoptively transferred into RIP-mOVA mice before the recipients were immunized with LPS and N4 peptide. Data are from three independent experiments (n = 20 mice). ****P < 0.0001 by log-rank Mantel-Cox test. (B) CFSE dilution in naïve CD8 T cells in vivo. CFSE-labeled naïve OT-I CD8+ T cells isolated from Prkd2+/+ and Prkd2SSAA/SSAA mice were mixed at a 1:1 ratio and adoptively transferred into C57BL/6 recipients before the mice were immunized with LPS with or without N4 peptide. Representative histograms show flow cytometric analysis of CFSE dilution 2 days after immunization. Graphs show the percentages of cells in each division in each of three independent experiments. (C) OT-I CD8+ T cells from Prkd2+/+ and Prkd2SSAA/SSAA mice were mixed and adoptively transferred into C57BL/6 recipients 2 days before they were immunized with LPS and N4 peptide. Seven days after immunization, splenic cells were isolated and stimulated in vitro with N4 peptide for 4 hours. IFN-γ production in OT-I donor cells was assessed by intra-cellular flow cytometric analysis. Histogram compares IFN-γ production in Prkd2+/+ and Prkd2SSAA/SSAA cells and is representative of three experiments. Graph shows the percentage of Prkd2SSAA/SSAA cells positive for IFN-γ relative to Prkd2+/+ cells. Data are from three independent experiments, with three to five mice per experiment. ***P = 0.0002 by Wilcoxon signed rank nonparametric test. (D to F) Naïve OT-I CD8+ T cells from Prkd2+/+ and Prkd2SSAA/SSAA mice were mixed at a 1:1 ratio and adoptively transferred into C57BL/6 recipients before the mice were subjected to L. monocytogenes OVA infection. Seven days later, donor cells were analyzed by flow cytometry. (D) Dot plots show the percentages of OT-I donor cells among the total CD8 population in blood and spleen, as determined by staining with congenic markers. Graphs show the mean percentages ± SD of OT-I Prkd2SSAA/SSAA cells relative to the percentage of OT-I Prkd2+/+ cells. Data are pooled from two independent experiments, with three to six mice per experiment. **P = 0.002 and *P = 0.0156 by Wilcoxon signed rank non-parametric test. (E) Histograms show the relative abundances of CD62L, CD44, and KLRG1 in the indicated donor OT-I cells as determined by flow cytometry. Graphs show the mean percentages ± SD of OT-I Prkd2SSAA/SSAA cells that were positive for KLRG1+ relative to the percentage of OT-I Prkd2+/+ cells that were KLRG1+. Data are pooled data from two independent experiments, with three to six mice per experiment. ***P = 0.001 and *P = 0.0156 by Wilcoxon signed rank nonparametric test. (F) Mean percentages ± SD of OT-I donor cells that were positive for IFN-γ relative to the percentage of OT-I Prkd2+/+ cells that were positive for IFN-γ. Data are pooled from two independent experiments, with three mice per experiment. *P = 0.0156 by Wilcoxon signed rank nonparametric test.
The effectiveness of T cell–mediated immune responses is determined by the number of effector T cells that can be generated from the pool of naïve T cells. To determine whether the phosphorylation and activation of PKD2 by PKCs was important for the production of effector T cells in vivo, we adoptively cotransferred carboxyfluorescein diacetate succinimidyl ester (CFSE)–labeled OT-I Prkd2+/+ and OT-I Prkd2SSAA/SSAACD8+ T cells into C57BL/6 hosts, immunized the mice with LPS and N4 peptide, and then analyzed TCR-dependent cell division and effector T cell differentiation. The data from these experiments showed that the TCR-stimulated clonal expansion of OT-I CD8+ T cells deficient in PKD2 activity was reduced compared to that of OT-I Prkd2+/+ T cells (Fig. 5B). OT-I Prkd2SSAA/SSAA CD8+ T cells also showed a marked defect in their ability to produce IFN-γ in response to immunization with LPS and N4 peptide (Fig. 5C). Hence, these data suggest that PKC-mediated activation of PKD2 is required to generate IFN-γ–producing effector T cells in vivo.
In additional experiments, we adoptively cotransferred OT-I Prkd2+/+ and OT-I Prkd2SSAA/SSAA naïve CD8+ T cells into C57BL/6 hosts and then infected the mice with an attenuated strain of Listeria monocytogenes expressing OVA (rLMOVA) (23). At the peak of the effector phase (day 7 after infection), it was clear that the magnitude of the OT-I Prkd2SSAA/SSAA T cell effector response was greatly reduced compared to that of their wild-type OT-I counterparts. Thus, the number of OT-I Prkd2SSAA/SSAA T cells present in the blood and spleen of immunized mice was about 50% of that of the OT-I Prkd2+/+ T cells (Fig. 5D). The OT-I Prkd2SSAA/SSAA T cells that we detected at the peak of the effector phase had increased the amount of CD44 and decreased the amount of CD62L at the cell surface, indicating that they had received stimulation through their TCR (Fig. 5E). We also analyzed the abundance of killer cell lectin-like receptor G1 (KLRG1), a cell surface marker that is diagnostic of terminally differentiated effector CD8+ T cells (24). There was a statistically significant decrease in the number of KLRG1+ terminally differentiated effector T cells among the OT-I Prkd2SSAA/SSAA CD8+ T cells (Fig. 5E), suggesting that the CD8+ T cell effector response was impaired in the absence of the catalytic activity of PKD2. In addition, there were less OT-I Prkd2SSAA/SSAA cells among the IFN-γ–producing cells in response to Listeria infection (Fig. 5F). Collectively, these data suggest that PKD2 is required for the efficient production of antigen-specific CD8+ effector T cells in vivo.
DISCUSSION
We showed that the DAG-regulated serine and threonine kinase PKD2 is a key signal amplifier for the TCR in naïve CD8+ T cells. The TCR-mediated activation of PKD2 was a digital response in which the percentage of T cells that activated this kinase was determined by the quality and quantity of the TCR ligand. Naïve CD8+ T cells have, on average, 400,000 molecules of PKD2, and the recognition of very few pMHC complexes on the surface of APCs was sufficient to activate the total pool of PKD2 in a single T cell. The effect of increasing the density and potency of pMHC ligands was such that the percentage of T cells that exhibited PKD2 activation increased. However, for each T cell that responded to antigen, the entire cellular pool of PKD2 was activated. Thus, PKD2 acts as a sensitive digital amplifier of TCR engagement to enable a population of naïve T cells to match the production of inflammatory cytokines to the quality and quantity of pMHC ligands. Previous studies showed that the number of cytokine-secreting naïve CD8+ T cells is determined by the strength of TCR-pMHC interactions (20). Here, we identified PKD2 as a key PKC substrate in T cells that functions as an amplifier of the TCR signaling pathway to determine the ability of a naïve T cell to commit to cytokine production.
One of our main findings was the observation of how sensitive PKD2 activity was to TCR engagement with cognate ligands. As few as 6 ± 103 pMHC molecules were sufficient to stimulate activation of up to 500,000 PKD2 molecules in a single T cell. The molecular mechanism that enables PKD2 to act as a sensitive amplifier of TCR signaling resides in its mode of activation. PKD activation is initiated by the binding of DAG to its N-terminal regulatory domains, but it is completed and stabilized by the PKC-mediated phosphorylation of Ser707 and Ser711 in the catalytic domain of PKD. The sensitivity of the response reflects the high affinity of PKD2 for DAG, such that PKD2 is rapidly recruited to the plasma membrane when the T cell makes contact with peptide-loaded APCs (6). PKD2 is then phosphorylated and activated by PKCs, and the active enzyme immediately relocalizes into the cytosol (6). The rapid return of active PKD to the cytosol effectively clears the plasma membrane to enable further recruitment of inactive PKD2 molecules. TCR stimulation thus initiates a rapid and continuous process of membrane recruitment, phosphorylation, and activation of PKD2. An activated T cell can thus rapidly process and activate the entire cellular pool of PKD2. The rapid recycling of active PKD2 to the cytosol so that more PKD2 can be recruited to and activated at the membrane is a potential mechanism that enables PKD2 to become the signal amplifier: only a few active PKC molecules at the membrane can rapidly interact with and phosphorylate the entire cellular pool of PKD2. In this context, the importance of PKD2 as a PKC substrate is demonstrated herein: when PKD2 could not be phosphorylated by PKC, the CD8+ effector T cell responses were markedly impaired. The signal amplification mediated by PKD2 in the context of its role as a PKC substrate functions to translate the strength and duration of a TCR stimulus to determine the size of the T cell effector pool.
Another feature of the PKD2 pathway that we found is that it displays a digital, all-or-none activation response to TCR stimulation. The digital nature of TCR-mediated PKD2 activation is not explained by any autonomous self-amplifying mechanism. Hence, when the TCR was bypassed with phorbol esters to activate PKD2, cells displayed a graded response and not the digital behavior seen in response to TCR stimulation. Thus, the digital response was not PKD2-intrinsic but dependent on TCR signaling. In this regard, recent models argue that the digital nature of TCR signaling is TCR-proximal and occurs because TCRζ phosphorylation by the tyrosine kinase ZAP70 creates a digital switch (25). Moreover, we have observed that the recruitment of PKD2 to the plasma membrane, which is mediated by DAG binding and hence marks sites of DAG accumulation at the membrane, is a digital response. Antigen-activated T cells thus recruit either all or none of their PKD to the plasma membrane (6). In addition, the TCR-stimulated activation of nuclear factor κB (NF-κB), another classic DAG- and PKC-mediated T cell response, is digital (26). The digital nature of the TCR-dependent activation of PKD2 thus reflects the digital nature of the way in which the TCR controls the two key molecules required for PKD2 activation: DAG and PKC.
In conclusion, our study offers new insights into the regulation of the PKD family of kinases and illustrates that they can act as digital amplifiers of signaling networks controlled by DAG and PKCs. Our work highlights the importance of highly quantitative, single-cell analyses to understand how cells control the activity of PKDs. We focused here on PKD2 in T cells; however, PKD2 is a member of a family of three kinases (PKD1, PKD2, and PKD3), all of which are activated by the binding of DAG as well as PKC-dependent phosphorylation (27). PKD1 and PKD3 have equally crucial roles in other cell lineages. Indeed, PKD1 is essential for embryonic development (1). Our observations about PKD2 regulation in T cells may thus be generally relevant to the roles of PKDs in other cell systems.
MATERIALS AND METHODS
Mice
OT-I Prkd2−/− and OT-I Prkd2SSAA/SSAA mice were previously described (2, 3). OT-I transgene expression in these mice was assessed by flow cytometry, and at least 95% of CD8+ T cells were routinely positive for the OT-I TCR. RIP-mOVA mice were purchased from The Jackson Laboratory. C57BL/6 mice were purchased from Charles River. Mice were maintained in the Biological Resource Unit at the University of Dundee in compliance with UK Home Office Animals (Scientific Procedures) Act 1996 guidelines. The University Ethical Review Committee approved the procedures.
Cell preparation and culture
Lymph nodes from 10- to 14-week-old mice were harvested, pooled, disaggregated, and resuspended in RPMI 1640 medium containing l-glutamine with 10% (v/v) heat-inactivated fetal bovine serum (FBS), penicillin (50 U/ml), streptomycin (50 μg/ml, Invitrogen), and 50 μM 2-mercaptoethanol (Sigma). OT-I TCR transgenic lymph node cells (5 × 106 cells/ml) were activated with the different OVA-derived peptides (N4, SIINFEKL; Q4R7, SIIQFERL; or T4, SIITFEKL) or with PDBu (Calbiochem) for the indicated times and at the indicated concentrations. Where indicated, naïve OT-I CD8+ T cells were purified before activation by magnetic sorting (autoMACS, Miltenyi) with a CD8 T cell isolation kit (Miltenyi) according to the manufacturer’s instructions. To analyze IFN-γ production by flow cytometry, total lymph node cells or splenocytes were activated as indicated earlier for 2 hours at 37°C before GolgiPlug (BD Biosciences) was added to the cells and incubated for an additional 4 hours.
Western blotting analysis
Protein abundance and phosphorylation in purified naïve OT-I CD8+ T cells from Prkd2+/+ and Prkd2SSAA/SSAA mice were assessed with standard Western blotting protocols. Briefly, cell lysates were prepared on ice with radioimmunoprecipitation assay buffer [50 mM Hepes (pH 7.4), 150 mM NaCl, 1% (w/v) NP-40, 0.5% (w/v) deoxycholate, 0.1% (w/v) SDS, 10% (w/v) glycerol, 1 mM EDTA, 1 mM EGTA, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 10 mM sodium β-phosphoglycerate, 0.5 mM sodium orthovanadate, 5 mM N-ethylmaleimide, 1 mM TCEP solution] supplemented with protease inhibitors (Roche) and were centrifuged at 16,000g for 10 min at 4°C. Protein samples were resolved by 10% bis-tris SDS–polyacrylamide gel electrophoresis gels, transferred onto nitro-cellulose membranes (GE Healthcare), and blocked with 5% (w/v) nonfat dried skimmed milk powder in phosphate-buffered saline (PBS) containing 0.05% Tween 20. Blots were incubated with antibodies specific for total PKD2, phosphorylated PKD2 (pPKD2), total ERK1/2, and pERK1/2-Thr202/Tyr204 (Cell Signaling Technology) according to the manufacturer’s instructions. The generation and characterization of pan- and phospho-specific antibodies against PKD2 has been described previously (2, 28).
Flow cytometry
For cell surface staining, cells were washed once in staining solution (PBS, 0.5% FBS, 0.5% bovine serum albumin, and 0.01% sodium azide) and stained for 20 min at 4°C in the same solution with saturating concentrations of antibody according to the manufacturer’s instructions. Antibodies conjugated with fluorescein isothiocyanate (FITC), PE, allophycocyanin (APC), peridinin chlorophyll protein (PerCP)–Cy5.5, PE-Cy7, Horizon V450, and Horizon PE-CF594 were purchased from BD Biosciences, and eFluor780-conjugated antibodies were obtained from eBioscience. Cells were stained for the surface expression of the indicated markers with antibodies against the following targets (clone numbers are in parentheses): CD8 (53-6.7), CD4 (RM4-5), CD62L (MEL-14), CD45.1 (A20), Vα2 (B20.1), and TCRβ (H57-597), which were obtained from BD Biosciences, and CD44 (IM7), CD45.2 (104), KLRG1 (2F1), and H-2Kb-SIINFEKL (25-D1.16), which were obtained from eBioscience. Data were acquired on either FACSCalibur or LSR Fortessa flow cytometers with DIVA software (BD Biosciences) and were analyzed with FlowJo software (Tree Star). Viable cells were gated according to their forward- and side-scatter profiles. Naïve OT-I CD8+ T cells were electronically gated as CD8+CD44low. Polyclonal, non-TCR transgenic naïve CD8+ T cells were gated as TCRβ+CD4−CD8+. For thymocyte staining, DP cells were gated as CD4+CD8+, CD4 SP cells were gated as TCRβhiCD4+CD8−, and CD8 SP cells were gated as TCRβhiCD4−CD8+. Intracellular staining with anti-PKD2 and anti–pPKD2-Ser873 antisera was performed as previously described (3, 28). Cells were washed once in staining solution, fixed in IC Fixation buffer (eBioscience) for 30 min on ice, washed, and permeabilized with 90% methanol for 30 min on ice. Cells were then stained in staining solution with saturating concentrations of the indicated antibodies for 30 min at room temperature, washed twice, and stained with Alexa 647–conjugated anti-rabbit antibody (Jackson ImmunoResearch), CD8-FITC, and CD44-PE for 30 min at room temperature. The same fixation and permeabilization protocol was followed for intracellular staining with Alexa Fluor 488–conjugated anti-pERK1/2 Thr202/Tyr204 (Cell Signaling Technology), CD8-APC, and CD44-PE. Cells were then washed twice in staining solution and analyzed by flow cytometry. For intracellular staining with anti–IFN-γ antibody (clone XMG1.2, BD Biosciences), cells were fixed, permeabilized, and stained with IC Fixation and Permeabilization buffers (eBioscience), according to the manufacturer’s instructions, and analyzed by flow cytometry.
Quantitation of peptide presentation in RMA-S
RMA-S TAP-deficient T cell lymphoma cells (provided by S. Powis, School of Medicine, University of St Andrews, St Andrews, UK) were loaded with serial dilutions of stimulating peptides for 2 hours at 37°C in serum-supplemented RPMI 1640 medium. Cells were washed and then stained with PE-coupled anti–SIINFEKL-Kb antibody (25D1.16, eBioscience), whose fluorescence was calibrated with Quantibrite beads (BD Biosciences) according to the manufacturer’s instructions. Peptide-loaded RMA-S cells were mixed with total lymph node cell preparations from OT-I mice at a ratio of 1:1 and incubated at 37°C for 2 hours before being processed for intracellular staining with anti–pPKD2-Ser873 antisera.
PKD2 quantification by SRM
Purified naïve OT-I CD8+ T cells from pooled lymph nodes were lysed in 8 M urea and 100 mM tris-HCl (pH 8), and proteins were digested with trypsin to generate peptides. Total lysate samples were separated on a 75 μm × 15 cm reversed-phase column (Thermo Fisher) in a Dionex Ultimate 3000 Nano LC system (Dionex/Thermo Fisher) coupled to a QTRAP 5500 (AB SCIEX) mass spectrometer according to the SRM method. Data were analyzed with Skyline software (29). Initial characterization of PKD2 peptides was performed to identify the best SRM transitions through experimental data, and the PKD2654–664 peptide (FLITQILVALR) was selected as optimal for PKD2 quantification by SRM. A known concentration of heavy-labeled PKD2654–664 peptide (arginine, R, +10 daltons, 13C6/15N4; AQUA QuantPro, Thermo Scientific) was mixed with a trypsin digest of total lysate of a known number of naïve OT-I T cells. The ratio of peak intensities of six distinct transitions for light (endogenous PKD2) and heavy peptides was used to back-calculate the amount of PKD2 per cell. The quantification was performed in three biological replicates.
Real-time quantitative PCR
RNA extracted with an RNeasy Mini kit (Qiagen) was reverse-transcribed with a qScript cDNA synthesis kit (Quanta BioSciences) according to the manufacturer’s protocol. Quantitative PCR (qPCR) analysis was performed in a 96-well plate with an iQ SYBR Green–based detection on an iCycler (Bio-Rad Laboratories). The abundance of Hprt mRNA was used for normalization, and derived values were averaged. Primers were as follows: Hprt, 5′-TGATCAGTCAACGGGGGACA-3′ (forward) and 5′-TTCGAGAGGTCCTTTTCACCA-3′ (reverse); Ifng, 5′-TTACTGCCACGGCACAGTC-3′ (forward) and 5′-AGATAATCTGGCTCTGCAGG-3′ (reverse).
ChIP assays
Purified naïve OT-I CD8+ T cells from pooled lymph nodes of Prkd2+/+ and Prkd2−/− mice were activated for 6 hours with N4 peptide and then fixed in 0.8% formaldehyde for 10 min at room temperature. Further cross-linking was prevented by adding glycine to a final concentration of 125 mM. Nuclear isolation was performed by lysis in hypotonic buffer [10 mM tris-HCl (pH 7.5), 10 mM EDTA, 0.5 mM EGTA, 0.5% NP-40, supplemented with protease inhibitors] for 30 to 60 min on ice and then centrifugation at 5000g for 10 min. Nuclear pellets were washed once in cold washing buffer [10 mM tris-HCl (pH 7.5), 1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl, supplemented with protease inhibitors] and then resuspended in cold nuclear lysis buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 2 mM EDTA, supplemented with protease inhibitors]. Chromatin fragments of about 500 base pairs were obtained by sonicating the nuclear lysates in a Branson 250 sonifier (10 cycles/4 s at 28 Å), and chromatin suspensions were centrifuged at 16,000g for 10 min at 4°C. Chromatin in the supernatant was immunoprecipitated overnight with anti–Pol II antibody (N-20, Santa Cruz Biotechnology) and coupled to protein G–agarose beads blocked with salmon sperm (Millipore) for the last 90 min of the incubation. Chromatin complexes were washed with (i) low-salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM tris-HCl (pH 8.0), 150 mM NaCl], (ii) high-salt buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM tris-HCl (pH 8.0), 500 mM NaCl], (iii) LiCl wash buffer [0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 20 mM tris-HCl (pH 8.0)], and (iv) TE buffer [10 mM tris-HCl (pH 8.0), 1 mM EDTA]. Chromatin was eluted with 10% Chelex for 15 min at 95°C, centrifuged at 16,000g, and analyzed by real-time PCR. The following primers for different regions of the Ifng locus were used: Cns1 (−6000), 5′-GTAATGCAATCACACCTTGGC-3′ (forward) and 5′-GCACTTTCTGTTGTACATGC-3′ (reverse); Prom1, 5′-GTGGATGAGAAATTCACATTAC-3′ (forward) and 5′-GTGCCATTCTTGTGGGATTC-3′ (reverse); Prom2, 5′-CACAAAACCATAGCTGTAATGC-3′ (forward) and 5′-GAAGGCTCCTCGGGATTACG-3′ (reverse); Prom3, 5′-GAGGAGCCTTCGACTAGGTA-3′ (forward) and 5′-CAAGTCAGAGGGTCCAAAGG-3′ (reverse); Codon1, 5′-CTCTGAGACAATGAACGCTAC-3′ (forward) and 5′-CAGTAACAGCCAGAAACAGC-3′ (reverse); Intron1, 5′-GTTGGTTGACCGTGGTTGATG-3′ (forward) and 5′-CCAGCAAATAGCAACGTAGC-3′ (reverse); Exon4, 5′-CAAGCATTCAATGAGCTCATC-3′ (forward) and 5′-CTTATTGGGACAATCTCTTCC-3′ (reverse); Cns2 (17.4 Kb), 5′-CCCCTGAAGTTCCTGTGAC-3′ (forward) and 5′-CACACACATCAGTTAAGAGTCC-3′ (reverse).
Adoptive transfers and the RIP-mOVA model
For in vivo proliferation analysis, purified OT-I cells expressing different congenic markers from OT-I Prkd2+/+ (CD45.1) and OT-I Prkd2SSAA/SSAA (CD45.1/CD45.2) mice were mixed at a 1:1 ratio and labeled with 5 μM CFSE for 10 min at 37°C. The mixed OT-I cells (5 × 106) were injected intraperitoneally into C57BL/6 (CD45.2) recipient mice. Two days later, the mice were immunized with 50 μg of N4 peptide (SIINFEKL) and 25 μg of LPS intraperitoneally. Two days after immunization, CFSE dilution was analyzed by flow cytometry. OT-I donor cells were electronically gated as CD8+Vα2+CD45.1+ (Prkd2+/+) and CD8+Vα2+CD45.1+CD45.2+ (Prkd2SSAA/SSAA). To assess IFN-γ production, splenocytes were isolated 7 days after immunization and were stimulated in vitro with 1 nM N4 peptide for 4 hours in the presence of GolgiPlug. A Wilcoxon signed rank non-parametric test was performed to determine the P values in these experiments with Prism 6 software (GraphPad). A similar immunization protocol was followed to induce diabetes in the RIP-mOVA model. Glucose concentration in the blood was monitored daily, and mice were scored as diabetic after two consecutive blood glucose measurements >14.3 mM. Survival curves were analyzed with the log-rank (Mantel-Cox) test in Prism 6 software.
Listeria infections
A total of 1 × 104 purified naïve OT-I CD8+ T cells from OT-I Prkd2+/+ (CD45.1/CD45.2) and OT-I Prkd2SSAA/SSAA (CD45.1) mice were mixed at a 1:1 ratio and injected intraperitoneally into C57BL/6 (CD45.2) recipient mice. Two days later, the mice were infected with 5 × 106 colony-forming units of attenuated ActA-deleted OVA-expressing L. monocytogenes [provided by H. Shen (23)] injected intravenously. Seven days after infection, blood and spleens were harvested, red blood cells were lysed, and lymphocytes were processed for flow cytometric analysis. OT-I donor cells were electronically gated as CD8+Va2+CD45.1+CD45.2+ (Prkd2+/+) and CD8+Va2+CD45.1+ (Prkd2SSAA/SSAA). To assess IFN-γ production, splenocytes were isolated and stimulated in vitro with 1 nM N4 peptide for 4 hours in the presence of GolgiPlug. Wilcoxon signed rank nonparametric test was performed to obtain P values with Prism 6 software.
Supplementary Material
Acknowledgments
We thank members of the Biological Services Unit, R. Clarke of the Flow Cytometry Facility, J. Rolf for assistance in the L. monocytogenes model, and all members of the D.A.C. laboratory and S. Matthews for critical reading of the manuscript.
Funding: This work was funded by Wellcome Trust Principal Research Fellowship grants 065975/Z/01/A and 097418/Z/11Z to D.A.C. M.T. and A.G.A. are funded by Medical Research Council UK and the pharmaceutical companies supporting the Division of Signal Transduction Therapy (AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Janssen Pharmaceutica, Merck KGaA, and Pfizer).
Footnotes
www.sciencesignaling.org/cgi/content/full/7/348/ra99/DC1
Fig. S1. Quantification of the number of pMHC molecules per RMA-S cell.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Fielitz J, Kim MS, Shelton JM, Qi X, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. Requirement of protein kinase D1 for pathological cardiac remodeling. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3059–3063. doi: 10.1073/pnas.0712265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthews SA, Navarro MN, Sinclair LV, Emslie E, Feijoo-Carnero C, Cantrell DA. Unique functions for protein kinase D1 and protein kinase D2 in mammalian cells. Biochem. J. 2010;432:153–163. doi: 10.1042/BJ20101188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarro MN, Sinclair LV, Feijoo-Carnero C, Clarke R, Matthews SA, Cantrell DA. Protein kinase D2 has a restricted but critical role in T-cell antigen receptor signalling in mature T-cells. Biochem. J. 2012;442:649–659. doi: 10.1042/BJ20111700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldron RT, Rey O, Iglesias T, Tugal T, Cantrell D, Rozengurt E. Activation loop Ser744 and Ser748 in protein kinase D are transphosphorylated in vivo. J. Biol. Chem. 2001;276:32606–32615. doi: 10.1074/jbc.M101648200. [DOI] [PubMed] [Google Scholar]
- 5.Rey O, Reeve JR, Zhukova E, Sinnett-Smith J, Rozengurt E. G protein-coupled receptor-mediated phosphorylation of the activation loop of protein kinase D: Dependence on plasma membrane translocation and protein kinase Cε. J. Biol. Chem. 2004;279:34361–34372. doi: 10.1074/jbc.M403265200. [DOI] [PubMed] [Google Scholar]
- 6.Spitaler M, Emslie E, Wood CD, Cantrell D. Diacylglycerol and protein kinase D localization during T lymphocyte activation. Immunity. 2006;24:535–546. doi: 10.1016/j.immuni.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Zehn D, King C, Bevan MJ, Palmer E. TCR signaling requirements for activating T cells and for generating memory. Cell. Mol. Life Sci. 2012;69:1565–1575. doi: 10.1007/s00018-012-0965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat. Immunol. 2004;5:524–530. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 9.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 10.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu. Rev. Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olenchock BA, Guo R, Carpenter JH, Jordan M, Topham MK, Koretzky GA, Zhong XP. Disruption of diacylglycerol metabolism impairs the induction of T cell anergy. Nat. Immunol. 2006;7:1174–1181. doi: 10.1038/ni1400. [DOI] [PubMed] [Google Scholar]
- 12.Zhong XP, Hainey EA, Olenchock BA, Jordan MS, Maltzman JS, Nichols KE, Shen H, Koretzky GA. Enhanced T cell responses due to diacylglycerol kinase z deficiency. Nat. Immunol. 2003;4:882–890. doi: 10.1038/ni958. [DOI] [PubMed] [Google Scholar]
- 13.Valverde AM, Sinnett-Smith J, Van Lint J, Rozengurt E. Molecular cloning and characterization of protein kinase D: A target for diacylglycerol and phorbol esters with a distinctive catalytic domain. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8572–8576. doi: 10.1073/pnas.91.18.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Brameshuber M, Zeng X, Xie J, Li QJ, Chien YH, Valitutti S, Davis MM. A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4+ T cells. Immunity. 2013;39:846–857. doi: 10.1016/j.immuni.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tkach K, Altan-Bonnet G. T cell responses to antigen: Hasty proposals resolved through long engagements. Curr. Opin. Immunol. 2013;25:120–125. doi: 10.1016/j.coi.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLOS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize Ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Holländer GA, Gascoigne NRJ, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 19.King CG, Koehli S, Hausmann B, Schmaler M, Zehn D, Palmer E. T cell affinity regulates asymmetric division, effector cell differentiation, and tissue pathology. Immunity. 2012;37:709–720. doi: 10.1016/j.immuni.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 22.Behrens GMN, Li M, Davey GM, Allison J, Flavell RA, Carbone FR, Heath WR. Helper requirements for generation of effector CTL to islet beta cell antigens. J. Immunol. 2004;172:5420–5426. doi: 10.4049/jimmunol.172.9.5420. [DOI] [PubMed] [Google Scholar]
- 23.Pearce EL, Shen H. Generation of CD8 T cell memory is regulated by IL-12. J. Immunol. 2007;179:2074–2081. doi: 10.4049/jimmunol.179.4.2074. [DOI] [PubMed] [Google Scholar]
- 24.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay H, Cordoba SP, Maini PK, van der Merwe PA, Dushek O. Systems model of T cell receptor proximal signaling reveals emergent ultrasensitivity. PLOS Comput. Biol. 2013;9:e1003004. doi: 10.1371/journal.pcbi.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kingeter LM, Paul S, Maynard SK, Cartwright NG, Schaefer BC. Cutting edge: TCR ligation triggers digital activation of NF-κB. J. Immunol. 2010;185:4520–4524. doi: 10.4049/jimmunol.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozengurt E. Protein kinase D signaling: Multiple biological functions in health and disease. Physiology. 2011;26:23–33. doi: 10.1152/physiol.00037.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews SA, Rozengurt E, Cantrell D. Characterization of serine 916 as an in vivo autophosphorylation site for protein kinase D/Protein kinase Cμ. J. Biol. Chem. 1999;274:26543–26549. doi: 10.1074/jbc.274.37.26543. [DOI] [PubMed] [Google Scholar]
- 29.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.