Abstract
Fragile-X syndrome is the most commonly inherited cause of autism and mental disabilities. The Fmr1 (Fragile-X Mental Retardation 1) gene is essential in humans and Drosophila for the maintenance of neural stem cells, and Fmr1 loss results in neurological and reproductive developmental defects in humans and flies. FMRP (Fragile-X Mental Retardation Protein) is a nucleo-cytoplasmic shuttling protein, involved in mRNA silencing and translational repression. Both Zfrp8 and Fmr1 have essential functions in the Drosophila ovary. In this study, we identified FMRP, Nufip (Nuclear Fragile-X Mental Retardation Protein-interacting Protein) and Tral (Trailer Hitch) as components of a Zfrp8 protein complex. We show that Zfrp8 is required in the nucleus, and controls localization of FMRP in the cytoplasm. In addition, we demonstrate that Zfrp8 genetically interacts with Fmr1 and tral in an antagonistic manner. Zfrp8 and FMRP both control heterochromatin packaging, also in opposite ways. We propose that Zfrp8 functions as a chaperone, controlling protein complexes involved in RNA processing in the nucleus.
Keywords: Zfrp8, FMRP, Trailer Hitch, translational repression
Introduction
Stem cell maintenance is essential for the generation of cells with high rates of renewal, such as blood and intestinal cells, and for the regeneration of many organs such as the brain and skin. We have previously shown that Zfrp8 is essential for maintaining hematopoietic, follicle, and germline stem cells (GSCs) in Drosophila melanogaster (Minakhina et al., 2014; Minakhina and Steward, 2010). Knockdown (KD) of Zfrp8 in GSCs results in the loss of stem cell self-renewal, followed by the eventual loss of all germline cells (Minakhina et al., 2014). Similarly in vertebrates, the Zfrp8 homolog, Pdcd2, is essential for embryonic stem cell maintenance and the growth of mouse embryonic fibroblasts; Pdcd2 mouse embryos die before implantation (Granier et al., 2014; Mu et al., 2010). PDCD2 is abundantly expressed and essential in highly proliferative cells including cultured cells and clinical isolates obtained from patients with hematologic malignancies (Barboza et al., 2013). The function of Zfrp8 and PDCD2 is highly conserved, as expression of transgenic PDCD2 is sufficient to rescue Zfrp8 phenotypes (Minakhina et al., 2014). Zfrp8 directly binds to Ribosomal Protein 2 (RpS2), a component of the small ribosomal subunit (40S), controls its stability and localization, and hence RNA processing (personal communication with Svetlana Minakhina). Zfrp8 also interacts with the piRNA pathway, which is conserved throughout all metazoans and is also essential for the maintenance of GSCs (Cox et al., 1998).
The piRNA pathway functions in maintaining heterochromatin stability and regulating the expression levels of retrotransposons. Both processes are thought to occur through piRNA targeting of chromatin modifying factors to the DNA. Guided by piRNAs, the piRNA pathway protein Piwi and associated proteins can set repressive epigenetic modifications to block transcription of nearby genes (Klenov et al., 2007; Le Thomas et al., 2013). Levels of transposon transcripts are also controlled by cytoplasmic PIWI-piRNA complexes, which can bind complementary mRNAs and mark them for translational repression and degradation (Lim et al., 2009; Rouget et al., 2010).
Fragile-X Mental Retardation Protein (FMRP) functions as a translational repressor involved in RNA silencing [reviewed in (Pimental and Tiossi, 2014)]. FMRP is a Piwi interactor and part of the piRNA pathway (Bozzetti et al., 2015; Megosh et al., 2006). FMRP-deficient animals display phenotypes similar to piRNA pathway mutants including genomic instability and de-repression of retrotransposons (Bozzetti et al., 2015; Deshpande et al., 2006). While FMRP is predominantly localized within the cytoplasm, FMRP complexes have also been demonstrated within the nucleus. In Xenopus, FMRP has been shown to bind target mRNAs co-transcriptionally in the nucleus (Kim et al., 2009). Like Zfrp8, FMRP has been shown to bind ribosomal proteins prior to nuclear export (Chen et al., 2014; Taha et al., 2014). In the cytoplasm, the FMRP-containing RNP complex controls mRNAs stability, localization, and miRNA-dependent repression (Chen et al., 2014; Napoli et al., 2008). FMRP mRNA targets are not well defined, as different studies show low overlap of putative targets in neuronal tissues (Brown et al., 2001; Chen et al., 2003; Darnell et al., 2001; Darnell et al., 2011; Miyashiro et al., 2003).
In Drosophila, FMRP is required to maintain GSCs, and loss of Fmr1 is associated with infertility and developmental defects in oogenesis and neural development (Callan et al., 2010; Costa et al., 2005; Wang et al., 2008; Yang et al., 2007). Fmr1, the gene encoding FMRP, is essential in both vertebrates and Drosophila for the maintenance of neural stem cells (NSCs) (Callan et al., 2010; Luo et al., 2010; Tervonen et al., 2010). In humans, loss of FMRP is associated with Fragile X-associated disorders, which cover a spectrum of mental, motor, and reproductive disabilities [reviewed in (Kidd et al., 2014; Santos et al., 2014; Sherman et al., 2014)]. Fragile X-associated disorders are the most commonly inherited cause of mental disabilities and autism (Hagerman, 2008). In vertebrates, FMRP physically interacts in the nucleus with NUFIP1 (Nuclear FMRP-Interacting Protein 1), a nucleo-cytoplasmic shuttling protein involved in ribonucleoprotein (RNP) complex formation (Bardoni et al., 2003; Boulon et al., 2008; McKeegan et al., 2009; Rothe et al., 2014). NUFIP1 is found in the nucleus in proximity to nascent RNA, and in the cytoplasm associated with ribosomes (Bardoni et al., 2003). In the cytoplasm, FMRP co-localizes and associates with Trailer Hitch (Tral) to form a translational repressor complex (Barbee et al., 2006). The Tral complex contains a number of translational repressor proteins, which together control the initiation of translation and the stability of mRNAs, such as gurken (grk) (Barbee et al., 2006; Jeske et al., 2011; Rouget et al., 2010; Wilhelm et al., 2005). In Drosophila, loss of Tral causes ovary phenotypes similar to piRNA pathway mutants, including oocyte polarity defects and transposon activation (Kugler et al., 2009; Liu et al., 2011; Snee and Macdonald, 2009).
In this study we identified Zfrp8 interactors by performing a yeast-two hybrid screen, and also by analyzing the components of the Zfrp8 complex by mass spectrometry. The nature of the proteins in the Zfrp8 complex indicates that it is involved in mRNA metabolism and translational regulation. We found that Zfrp8, Nufip, FMRP, and Tral are all part of the complex and we show that Zfrp8 interacts antagonistically with Fmr1 and tral, suppressing their oogenesis defects. Furthermore, we determined that Zfrp8 is required within the nucleus, and controls FMRP localization within the cytoplasm. We further confirm that FMRP functions in heterochromatin silencing and that Zfrp8 is required in the same process, but has an opposite function of FMRP. We propose that Zfrp8 functions as a chaperone of the FMRP' containing RNP translational repression complex and controls the temporal and spatial activity of this complex.
Materials and Methods
Fly lines and genetic interactions
Germline expressing VALIUM22 constructs were used for RNAi experiments. UAS-Zfrp8 RNAi (TRiP# GL00541, BDSC# 36581) and UAS-tral RNAi lines (TRiP# GL00680, BDSC# 38908) were obtained from the TRiP at Harvard Medical School, Boston, MA, USA. The hsp70-Gal4 driver (P[GAL4-Hsp70.PB]89-2-1), Df(3R)Exel6265, and PEV reporters (P[hsp26-pt-T, hsp-70w+]118E10-C4, P[hsp26-pt-T, hsp-70w+]118E15-T4) were obtained from the Bloomington Stock Center. The nos-Gal4 driver (P[GAL4::VP16-nos.UTR]) was obtained from T. Schupbach (Princeton, NJ, USA). The Fmr13 line was a gift from T.A. Jongens (Philadelphia, PA, USA) and the Fmr1Δ50M line was from D.C. Zarnescu (Tucson, AZ, USA). The Df(3R)Exel6265 line was obtained from the Bloomington Stock Center (BDSC# 7732). The Tral-GFP reporter protein trap line (P[w+mC=PTT-un1]G00140) was received from L. Cooley and the FlyTrap Project (New Haven, CT, USA) (Morin et al., 2001). In all experiments w118 flies were used as wild type controls.
For egg phenotype and fertility assays, 1 day-old females and males were set up on egglay plates and were changed daily over 5 days. The number of eggs laid was counted when the plate was changed and egg phenotypes and fertility rates were assessed 2 days later. Ventralization phenotypes were scored as previously described (Li et al., 2014).
Zfrp8 constructs
The Zfrp8 coding region was amplified by RT-PCR and cloned into a Gateway pENTR4 (Life Technologies) vector. Zfrp8 deletion constructs were created via PCR site-directed mutagenesis. The Zfrp8 NLS deletion construct removes putative NLS sequences at residues 100–106 and 246–263. The Zfrp8 NES deletion construct removes a putative NES sequence at residues 304–317. Deletion constructs were then cloned into pUAS-TAP-mCherryW-attB vector for injections (Hudson and Cooley, 2010).
For targeted Zfrp8 constructs, A GFP coding sequence was then subcloned at the 5’ end of Zfrp8 to create pENTR4-GFP-Zfrp8 (Gateway). To create membrane-localized CD8-GFPZfrp8, transgenic mouse CD8a was amplified from y1w*; P[UAS-mCD8::GFP.L]LL5 (BDSC #5137) and subcloned at the 5’ end of the GFP coding sequence (Lee and Luo, 1999). Nuclear-localized GFP-NLS-Zfrp8 and cytoplasmic-localized GFP-NES-Zfrp8 constructs were created by amplifying pENTR-GFP-Zfrp8 via circular PCR, using primers with extended 5’ NLS and NES coding sequences, respectively. The NLS sequence encodes the SV40 Large T-antigen monopartite NLS, PKKKRKV (Kalderon et al., 1984). The NES sequence encodes the HIV-1 Rev NES, LPPLERLTLD (Fischer et al., 1995). The inserts were transferred into pUASg-attB plasmids using the Invitrogen Gateway Cloning System (Bischof et al., 2013). Transgenic fly lines were created via PhiC31 integrase-mediated transgenesis inserted into the attP2 landing site (Groth et al., 2004) by Genetics Services, Inc. at Cambridge, MA, USA.
For targeted rescue experiments, transgenic Zfrp8 lines were crossed to hsp70-Gal4 in a Zfrp8 mutant background and raised at 25°C. Viability was calculated by comparing the number of actual eclosed adults to total expected adults. For mutational analysis and genetic interaction experiments, crosses were raised until eclosion at 29°C, and subsequently maintained as adults at 25°C until examination.
Position effect variegation
Ethanol-based pigment extraction and quantification was performed as described in Sun et al. (2004) with minor modifications. Flies were homogenized in 250ul pigment assay buffer, followed by incubation at room temperature for 1 hour for pigment extraction. A final volume of 200 ul of pigment extract was used to read OD at 480 nm. For each assay, data from 3 samples (each sample made up of twenty 3 day old flies, randomly picked from the population) were collected.
Protein purification and mass spectrometry
To isolate the Zfrp8 protein complex, tandem affinity purification (TAP) was done as described in (Burckstummer et al., 2006; Kyriakakis et al., 2008; Veraksa et al., 2005). Zfrp8 was cloned into pUAST-NTAP (Veraksa et al., 2005). Transgenic flies carrying pUAST-NTAP-Zfrp8 were generated using standard methods (Brand and Perrimon, 1993). Expression of NTAP-Zfrp8 under the da-Gal4 driver was sufficient to rescue Zfrp8 lethality and sterility. Extracts of da-Gal4/UAS-NTAP-Zfrp8 and w118 (control) 0–12 hr embryos were used for two step affinity purification. Proteins were separated by SDS-PAGE and bands visualized by Coomassie staining. To eliminate the contribution from IgG and Zfrp8 itself, fragments from 60–200 kD and from 15–35 kD were cut from the gel and analyzed by the Biological Mass Spectrometry Facility of the University of Medicine and Dentistry of New Jersey–Rutgers for LC-MS/MS analysis. Positive proteins were represented by ≥5 peptides in Zfrp8 fractions and also ≤1 peptides in the vector only control.
Nufip was cloned into the pMK33-NTAP vector (Veraksa et al., 2005). Drosophila S2 cells were transfected with either pMK33-NTAP-Nufip or the vector alone (as a control). After transfection and selection of stable cell lines, cells were grown for 8 days at 18°C before lysis and tandem affinity purification. Input and immunoprecipitation fractions were probed with anti-FMRP and anti-Zfrp8 antibodies.
For co-immunoprecipitation experiments, human NUFIP1 was cloned into a pCDNA 3xFLAG vector. After transfection of HEK293 cells with this construct or the empty vector, cells were grown for 72 hours at 37°C before lysis and immunoprecipitation using a Sigma-Aldrich FLAG Immunoprecipitation Kit as described in the manufacturer’s instructions. Extracts from cells transfected with the empty FLAG tag vector were used as negative controls. Input and immunoprecipitation fractions were probed on western blot with anti-FLAG and anti-PDCD2 antibodies.
Yeast two-hybrid screen
The PDCD2 yeast two-hybrid screen was performed using the Matchmaker Gold Protocol Yeast-Two Hybrid System (Clontech #630489). Matchmaker uses 4 different reporters each under the control of a distinct and separate cell cycle-responsive promoter, M1-expressing AUR1-C (Aureobasidin A/AbA resistance), M1-expressing MEL1 (α-galactosidase), G1-expressing HIS3 (histidine biosynthesis) and G2-expressing ADE2 (adenine biosynthesis). For bait, full-length human PDCD2 was cloned into the pGBKT7 vector. Expression of PDCD2 in yeast cells was confirmed by Western blotting using anti-PDCD2, and tested negative for auto-activation and toxicity. The pGBKT7-PDCD2 construct was then mated to a Mate and Plate normalized mouse embryonic stem cell library (Clontech #630484). Mated yeast culture was plated onto low-stringency plates containing minimal, synthetically defined (SD) -Leu/-Trp/X-α-Gal/AbA. Positive colonies were confirmed on high-stringency plates containing (SD) -Ade/-His/-Leu/-Trp/X-α-Gal/AbA. Positive plasmids were then isolated and retested with pGBKT7-PDCD2 on high stringency plates for final confirmation. Retested positives were then sequenced to identify PDCD2 interactors.
Protein prediction software
Ortholog prediction was completed using DIOPT- DRSC Integrative Ortholog Prediction Tool (Hu et al., 2011). Prediction of Zfrp8 nuclear localization signals was completed using cNLS Mapper (Kosugi et al., 2009). Prediction of Zfrp8 nuclear export signal was completed using ExPASy NetNES (la Cour et al., 2004).
Immunostaining and microscopy
For immunostaining, ovaries were dissected from either virgins (<12 hours) or at 7 days after eclosion, as indicated. Lymph glands were dissected from third instar larvae. For each immunostaining experiment, a minimum of 15 samples were analyzed.
Rabbit anti-Zfrp8 antibody was used at 1:2500 (Minakhina et al., 2014). Monoclonal mouse anti-FMRP 5B6 (DSHB, University of Iowa) was used at 1:1000. Monoclonal mouse anti-FLAG M2 (Sigma) was used at 1:1000. Polyclonal rabbit anti-PDCD2 (1:1000) was a gift from P.A. Sharp (Cambridge, MA, USA) (Scarr and Sharp, 2002). Alexa Fluor 488 Phalloidin (Invitrogen) and secondary antibodies (Jackson Laboratories) were used at 1:300. Hoechst 33258 (1:5000) was used to stain DNA. Ovary immunostaining images were captured using a Leica TCS SP5 laser scanning confocal microscope (at 63× oil), analyzed with Leica Microsystems software and processed using Adobe Photoshop. Egg phenotype images were captured using a Zeiss SteREO Discovery. V8 stereomicroscope (at 5×), analyzed with ProgRes Mac CapturePro 2.6 software and processed using Adobe Photoshop.
Quantitative RT-PCR
Quantitative RT-PCR was performed as described in the manufacturer instructions using the QuantiTect SYBR Green kit (Qiagen), Smart Cycler II (Cepheid) and the relative standard curve method. RNA was isolated from 10–20 virgin ovaries (<12 hours old) using Qiagen RNeasy Plus Mini kits. Confirming knockdown in the tral RNAi line, we quantified tral expression in nos-Gal4/UAS-tral RNAi at 0.320 ±0.045 s.d., compared to nos-Gal4/+ expression at 1.043 ±0.053 s.d. from whole ovaries. w118 control ovaries used as the baseline (equal to 1). Transcript levels were normalized to those of Gapdh1. A minimum of two biological and two technical replicates were performed for each genotype. Primers used for tral qPCR were: AAATGCCACAACCGCGAC, AAAGTGGCTTTCCACTGGC
Results and Discussion
Nufip and FMRP are components of the Zfrp8 protein complex
Zfrp8 is essential for stem cell maintenance, but its molecular functions have not yet been clearly defined (Minakhina et al., 2014; Minakhina and Steward, 2010). In order to address this question we used two distinct approaches. We performed a yeast-two hybrid screen to identify direct interactors of Zfrp8 and we also characterized the components of the Zfrp8 complex by mass spectrometry.
Because of the high sequence and functional conservation of Zfrp8 (flies) and PDCD2 (mammals) (Minakhina et al., 2014), and because no stem cell-derived cDNA library exists in Drosophila, we decided to screen a mouse embryonic stem cell cDNA library using mammalian PDCD2 as bait (see Materials and Methods). We isolated 46 initial positives, and identified 19 potential interactors after re-testing of the positives (Supplemental Table 1).
In order to purify the Zfrp8 protein complex we established a transgenic line expressing NTAP-tagged Zfrp8 under the control of the general da-Gal4 (daughterless) driver. Two-step tandem affinity purification was performed on embryonic extracts and the purified proteins were separated by SDS-PAGE electrophoresis. The proteins were eluted and analyzed by mass spectrometry (see Materials and Methods). Thirty proteins were identified as part of the Zfrp8 complex. The threshold for interactors was set to at least 5× peptide enrichment in Zfrp8 over vector control fractions (Table 1). Eighteen of the proteins are predicted to function in ribosomal assembly or translational regulation, strongly suggestive of a function of Zfrp8 in mRNA processing (i.e. translation, localization, and stability). In the complex we found six ribosomal subunits (five 40S subunits and one 60S subunit); EF2 and eIF-4a, which are required for translation initiation and elongation; and FMRP, Tral and Glorund which function in mRNA transport and translational repression. While Zfrp8 interacts with several ribosomal proteins it does not appear to be part of the ribosome itself (Marygold et al., 2007).
Table 1.
Zfrp8 interactors identified by TAP purification followed by mass spectrometry
| Symbol | Protein | Human Ortholog | # Peptides Matched* | Estimated Mass (kD) | Molecular Function |
|---|---|---|---|---|---|
| Act42A | Actin 42A | ACTB/G1 | 33 | 41.8 | cytoskeletal protein |
| AP-2α | AP-2 complex subunit alpha | AP2A2 | 10 | 105.6 | endocytosis |
| CkIα | Casein kinase I isoform alpha | CSNK1A1 | 21 | 39.5 | protein kinase |
| Ef1γ | Elongation factor 1-gamma | EEF1G | 23 | 49.0 | regulation of organelle transport (Serpinskaya et al., 2013) |
| EF2 | Elongation factor 2 | EEF2 | 18 | 94.5 | translation elongation factor |
| eIF-4a | Eukaryotic initiation factor 4A | EIF4A1/2 | 24 | 45.9 | translation initiation factor |
| FK506-bp1 | 39 kDa FK506-binding nuclear protein | 12 | 39.3 | prolyl isomerase | |
| FMRP | Fragile-X mental retardation protein | FMR1 | 8 | 72.0 | Translational regulation, Rm62-interactor (Zhang et al., 2001; Ishizuka et al., 2002) |
| Glo | Glorund | HNRNPH1 | 39 | 61.4 | Gurken mRNA localization and translational repression (Kalifa et al., 2009) |
| Hrb27C | Heterogeneous nuclear ribonucleoprotein 27C | DAZAP1 | 26 | 44.8 | Gurken mRNA localization (Goodrich et al., 2004) |
| Hrb87F | Heterogeneous nuclear ribonucleoprotein 87F | HNRNPA2B1 | 50 | 39.5 | ribonucleoprotein, NonA-interactor (Reim et al., 1999) |
| Hsc70-4 | Heat shock 70 kDa protein cognate 4 | 23 | 71.1 | chaperone protein | |
| Hsp83 | Heat shock protein 83 | HSP90AA1/B1 | 8 | 81.9 | chaperone protein, regulation of piRNA pathway (Gangaraju et al., 2011) |
| Map205 | 205 kDa microtubule-associated protein | 25 | 126.7 | cytoskeletal protein | |
| Nep1 | Neprilysin 1 | MME/L1 | 31 | 96.5 | peptidase |
| NonA | Protein no-on-transient A | SFPQ | 163 | 77.0 | RNP nucleo-cytoplasmic shuttling (Kozlova et al., 2006) |
| Nop60B | Nucleolar protein at 60B | DKC1 | 54 | 56.8 | ribosomal RNA processing (Giordano et al., 1999) |
| Noppl40 | Noppl40 | NOLC1 | 34 | 70.5 | ribosome assembly and repression of transposon expression (He et al., 2014) |
| Pug | Pugilist | MTHFD1 | 20 | 103.5 | tetrahydrofolate conversion |
| Rm62 | Rm62 | DDX17 | 68 | 50.0 | RNA helicase, FMRP-interactor (Ishizuka et al., 2002) |
| RpL7A | Ribosomal protein L7A | RPL7A | 7 | 30.7 | ribosomal protein |
| RpS2 | Ribosomal protein S2 | RPS2 | 224 | 28.9 | ribosomal protein (Cramton and Laski, 1994) |
| RpS3 | Ribosomal protein S3 | RPS3 | 15 | 27.5 | ribosomal protein |
| RpS4 | Ribosomal protein S4 | RPS4 | 32 | 29.1 | ribosomal protein |
| RpS5a | 40S ribosomal protein S5a | RPS5 | 11 | 25.4 | ribosomal protein |
| RpS7 | Ribosomal protein S7 | RPS7 | 18 | 22.2 | ribosomal protein |
| Top2 | DNA topoisomerase 2 | TOP2A/B | 59 | 164.4 | topoisomerase |
| Tral | Trailer hitch | LSM14A/B | 80 | 69.3 | Secretory pathway regulation; Repression of transposon expression, FMRP-interactor (Kugler et al., 2009; Liu et al., 2011) |
| αTub84B | Alpha-tubulin at 84B | TUBA3C/D | 5 | 49.9 | cytoskeletal protein |
| βTub56D | Beta-tubulin at 56D | TUBB4B | 8 | 51.3 | cytoskeletal protein |
For all proteins, ≤1 peptides were found in the vector-only TAP control
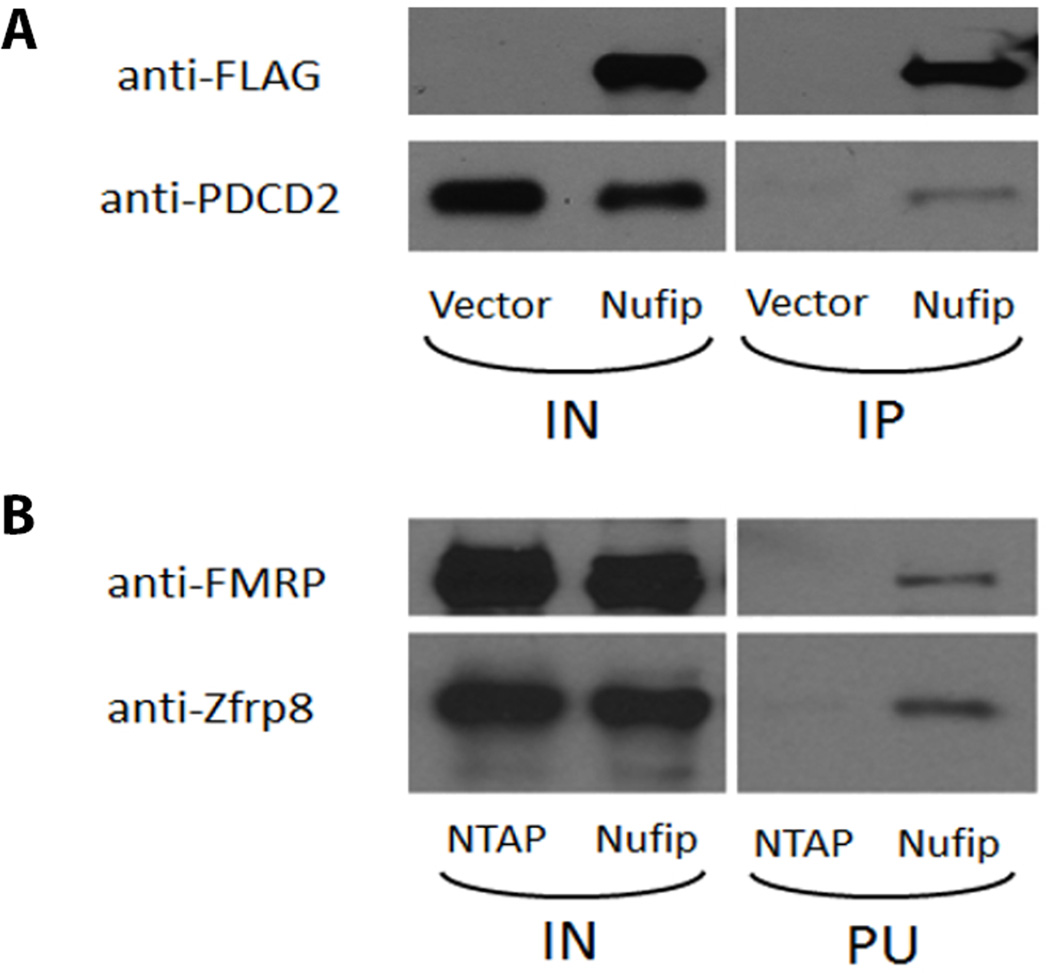
No overlapping interactors were found in our yeast-two hybrid screen and mass spectrometry assay. But interestingly, FMRP was identified as part of the Zfrp8 complex by mass spectrometry and NUFIP1 in our yeast-two hybrid assay. Most likely Nufip (estimated 57 kD) was not identified as part of the Zfrp8 complex in the TAP-purification approach, because we excluded proteins with similar size to tagged Zfrp8 (~55 kD) from the mass spectrometry analysis. To investigate whether these proteins could work together in the same molecular process we confirmed the interaction of both Zfrp8 and PDCD2 with Nufip (flies) and NUFIP1 (mammals) in tissue culture cells. Immunoprecipitation of human HEK293 cell extracts expressing FLAG-tagged NUFIP1 pulled down endogenous PDCD2 (Fig. 1A). We next examined whether this protein interaction also exists in Drosophila. We were able to co-purify endogenous Zfrp8 with NTAP-tagged Nufip from transfected S2 cells (Fig. 1B). We then performed an additional Western blot on the purified NTAP-Nufip isolate and could show that FMRP is present in the protein complex (Fig. 1B), indicating that Nufip physically interacts with both Zfrp8 and FMRP. Our results suggest that all three proteins function together in a molecular complex which regulates RNP processing/assembly and translation. Based on these results, and the requirement of both Zfrp8 and Fmr1 in stem cell maintenance, we decided to characterize the genetic interaction between these genes.
Figure 1. Nufip and FMRP are components of the Zfrp8 complex.
A. Western blot displaying the immunoprecipitation of human PDCD2 with FLAG-tagged NUFIP1 from HEK293 cell extracts. Negative control: extract from cells transfected with the empty FLAG-tag vector. B. NTAP-tagged Drosophila Nufip was expressed in S2 cells and purified. Protein complex is visualized on western blot with anti-FMRP and anti-Zfrp8 antibodies. Negative control: extract from cells transfected with the empty NTAP-tag vector. IN – total extract, IP immunoprecipitate, PU purified complex.
Loss of Zfrp8 suppresses Fmr1 infertility and ovary defects
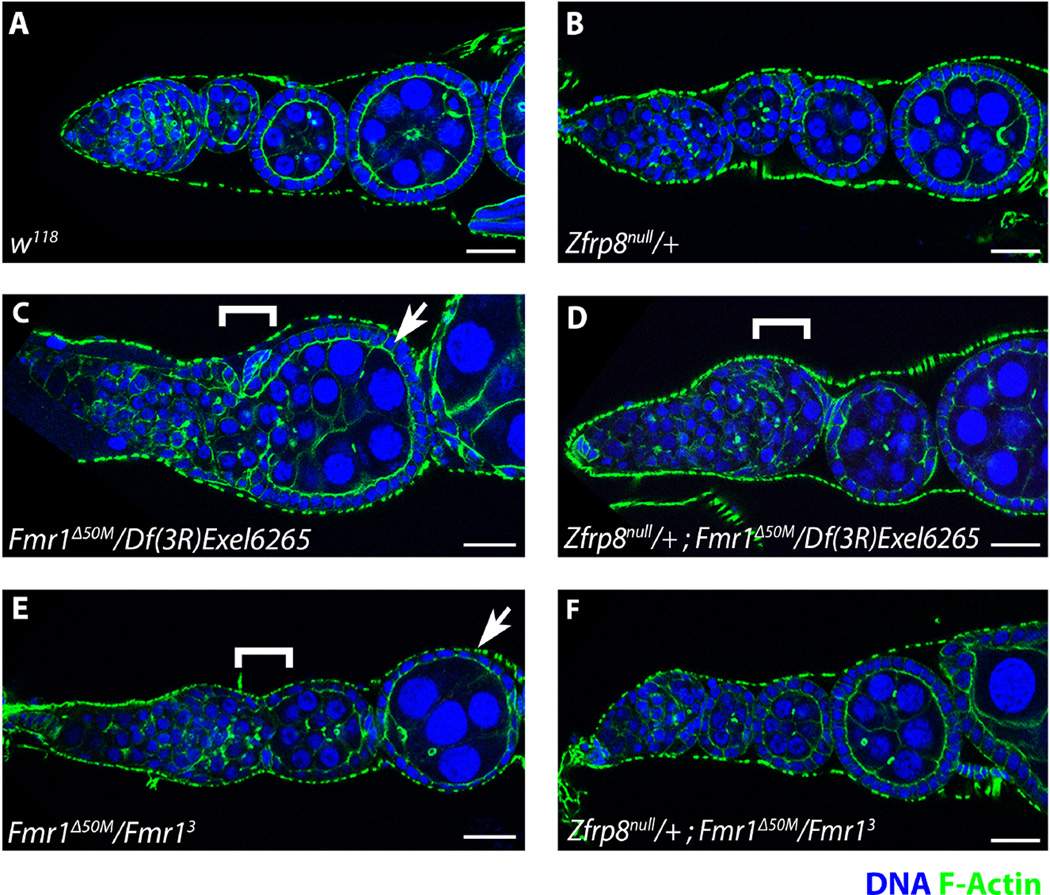
To further characterize the connection between the two genes, we examined whether the loss of Zfrp8 can modify oogenesis defects reported for Fmr1 females (Costa et al., 2005). Similar to what was previously reported, 100% of Fmr1Δ50M/Df(3R)Exel6265 (N = 26) and 80% of Fmr1Δ50M/Fmr13 (N = 22) ovaries displayed developmental defects (Costa et al., 2005). The ovarioles contained fused egg chambers (100%, 64% for each genotype, respectively; Fig.2C and 2E, bracket), aberrant nurse cell numbers (46%, 32% ; Fig.2C and 2E, arrow). We occasionally also observed egg chambers with oocyte misspecification/multiple oocytes (< 20%, not shown). Interestingly, the loss of one copy of Zfrp8 suppressed the majority of Fmr1 ovary defects, restoring cell division in the germline, as well as egg chamber morphology and separation (Fig. 2D, 2F). In Zfrp8/+; Fmr1Δ50M/Df(3R)6265 (N=20), fusion of the first egg chamber is still observed in most germaria, but despite this, oogenesis appears to proceed normally resulting in normal looking ovarioles (Fig. 2D, bracket). Zfrp8/+; Fmr1Δ50M/Fmr13 (N=33) ovaries appear almost completely normal even though these ovarioles contain no FMRP (Fig. 2F).
Figure 2. Loss of Zfrp8 suppresses Fmr1 infertility and ovary defects.
A. Wild-type ovariole comprised of a normal germarium and early egg chambers. B. Zfrp8null/+ heterozygote ovarioles do not display any morphological defects. C. Fmr1Δ50M/Df(3R)Exel6265 ovarioles displays a disorganized germarium, often fused to an egg chamber containing more than the normal 15 nurse cells (bracket and arrow). D. Zfrp8null/+; Fmr1Δ50M/Df(3R)Exel6265 ovarioles show suppression of the Fmr1Δ50M/Df(3R)Exel6265 morphological defects. While the first egg chamber often remains fused to the germarium (bracket), the later stages of oogenesis are normal. E. Fmr1Δ50M/Fmr13 ovariole containing a fused germarium-egg chamber (bracket) and an egg chamber with abnormal numbers of nurse cells (arrow). F. Zfrp8null/+ ; Fmr1Δ50M/Fmr13 exhibit morphologically normal ovarioles. DNA (blue); filamentous actin-phalloidin (green). Confocal sections are shown; scale bar: 10 uM.
The loss of Fmr1 has also been associated with a strong reduction in egg production (Bauer et al., 2008; Zhang et al., 2004). We found that similar to previous reports, Fmr1Δ50M/Df(3R)Exel6265 and Fmr1Δ50M/Fmr13 mutants display a strong reduction in fertility; females laid on average of 1 and 6 eggs/day, respectively, as compared to 18 eggs/day for wildtype flies (Table 2). The removal of one copy of Zfrp8 partially suppressed Fmr1 infertility and resulted in 8 eggs/day from Fmr1Δ50M/Df(3R)Exel6265 and 15 eggs/day from Fmr1Δ50M/Fmr13 females. These results demonstrate that Zfrp8 and Fmr1 affect the same process and that even though they are found in the same complex, have opposing functions.
Table 2.
Zfrp8 suppresses Fmr1 egg laying defects
| Genotype | Hatch Rate | Eggs Laid per Day |
Eggs Counted |
|---|---|---|---|
| w118 | 95.10% | 18.01 | n= 1801 |
| Fmr1Δ50M/+ | 91.68% | 16.84 | n= 317 |
| Df(3R)Exel6265/+ | 90.79% | 12.75 | n= 956 |
| Fmr1Δ50M/Df(3R)Exel6265 | 94.85% | 0.97 | n= 97 |
| Zfrp8null/+ ; Fmr1Δ50M/Df(3R)Exel6265 | 85.99% | 8.28 | n= 236 |
| Fmr1Δ50M/Fmr13 | 89.55% | 5.74 | n= 145 |
| Zfrp8null/+ ; Fmr1Δ50M/Fmr13 | 78.48% | 15.14 | n= 336 |
Zfrp8 is required for proper FMRP localization
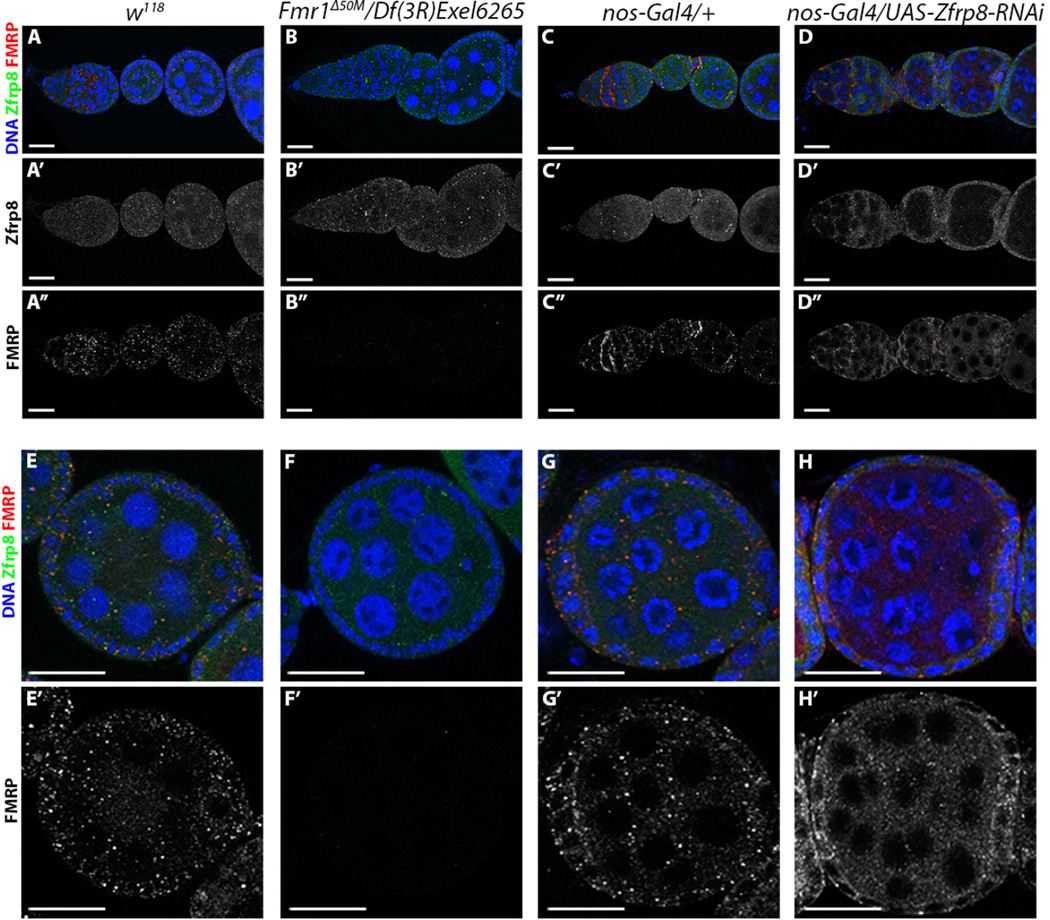
To investigate the nature of the Zfrp8 interaction with FMRP, we examined the localization of the proteins within the ovary. As we have shown previously, Zfrp8 displays ubiquitous distribution in all cells and cell compartments of the wild type ovary (Fig. 3A, A’). No significant changes in Zfrp8 localization or levels are visible in Fmr1 ovaries (Fig. 3B, B’). FMRP has a more varied distribution pattern, present in strong, cytoplasmic puncta in the cytoplasm of nurse cells and follicle cells (Fig. 3A, A”, E, E’), and also in high levels in the cytoplasm of the maturing oocyte (3E, E’). FMRP is also detectable in low levels in nurse cell nuclei at stage 8 egg chambers at an average of 9.76 puncta per nucleus (N=80). As expected, Fmr1 ovaries display no FMRP staining in either the cytoplasm or nucleus (Fig. 3B, B”, F, F’,).
Figure 3. Zfrp8 is required for proper FMRP localization.
A–A”, E–E’. In wild-type (w118) ovarioles FMRP is seen as puncta throughout the cytoplasm and at low levels in nurse cell nuclei of stage 8 egg chambers at an average of 9.76 puncta per nucleus (N=80), while Zfrp8 is uniformly distributed in the cytoplasm and nuclei. B–B”, F–F’. In Fmr1Δ50M/Df(3R)Exel6265 ovarioles FMRP protein is absent as expected both in the cytoplasm and in nuclei (N=81), while Zfrp8 expression and distribution is not significantly changed. C–C”, G–G’. Control nos-Gal4/+ ovarioles appear similar to wild-type, FMRP localizes to cytoplasmic foci;. D–D”, H–H’. Within Zfrp8 KD egg chambers FMRP localization to cytoplasmic puncta is disrupted and the protein is more uniformly distributed throughout the cytoplasm. Anti-Zfrp8 (green), anti-FMRP (red), DNA (blue). Confocal sections are shown; scale bar: 10 uM.
To determine whether Zfrp8 functions in FMRP regulation, we depleted Zfrp8 in the germline by expressing Zfrp8 RNAi under the control of the nos-Gal4 driver (Minakhina et al., 2014), and assessed changes in FMRP expression. In control nos-Gal4 ovaries, FMRP levels and distribution were similar to that in wild-type ovaries (Fig. 3C, C”, G, G’). However, in Zfrp8 KD ovaries, aberrant FMRP localization is observed in the germline; FMRP is more uniformly distributed throughout the cytoplasm and puncta are strongly diminished (Fig. 3D, D”, H, H’). Remaining FMRP puncta appear fragmented, reduced in intensity, size and number (~10% of wild-type; N=22 egg chambers counted). These results indicate a Zfrp8 requirement for proper FMRP localization to the cytoplasm. FMRP normally functions by shuttling mRNA cargo from the nucleus to the cytoplasm, where it represses the translation of bound mRNA. The observed change of FMRP localization in Zfrp8 KD ovaries therefore may indicate a regulatory function for Zfrp8 in the nuclear export and localization of FMRP.
Zfrp8 is required in the nucleus
Zfrp8 protein is present in both the cytoplasm and nucleus (Fig. 3A, A’) (Minakhina et al., 2014) and, as demonstrated above, controls the distribution of FMRP in the cytoplasm. We decided to investigate the cell compartment in which Zfrp8 is required, in order to elucidate how Zfrp8 regulates FMRP. To do so, we examined the capability of Zfrp8 deletion constructs to rescue mutant lethality. Expression of human PDCD2 cDNAs driven by the general driver da-Gal4 is fully capable of rescuing Zfrp8 lethality (Barboza et al., 2013; Minakhina et al., 2014). We created mutated Zfrp8 constructs, removing either the two putative NLSs or the putative NES domains. These proteins were expressed under the da-Gal4 driver, and while clearly overexpressed on Western blots, failed to rescue mutant lethality, suggesting that the three domains are essential for the function of the protein (not shown).
In an alternative approach, we assayed the function of Zfrp8 proteins targeted to a distinct cell compartment. We expressed four N-terminal GFP-tagged transgenic proteins encoding a wild-type Zfrp8, nuclear-localized NLS-Zfrp8, cytoplasmic-localized NES-Zfrp8, and cell membrane-localized CD8-GFP-Zfrp8. Transgenic Zfrp8 subcellular localization is visible when the proteins are strongly overexpressed (Supplemental Fig. 1A–D). When we expressed the transgenes at lower levels, similar to the endogenous levels, with the hsp70-Gal4 driver at 25°C, both wild-type and nuclear-localized Zfrp8 were able to rescue mutant lethality at similar rates, whereas the cytoplasmic- and membrane-localized proteins did not show rescue (Supplemental Table 2). These results show that Zfrp8 is required in the nucleus and suggest that like FMRP, Zfrp8 may function by shuttling between nuclear and cytoplasmic compartments.
Zfrp8 suppresses the tral oogenesis phenotypes
We have shown that FMRP and Zfrp8 are present in the same protein complex. In addition to FMRP, our mass spectrometry results have also identified other translational regulators, such as Tral. Tral has previously been shown to function in conjunction with FMRP to control the translation of mRNAs (Barbee et al., 2006).
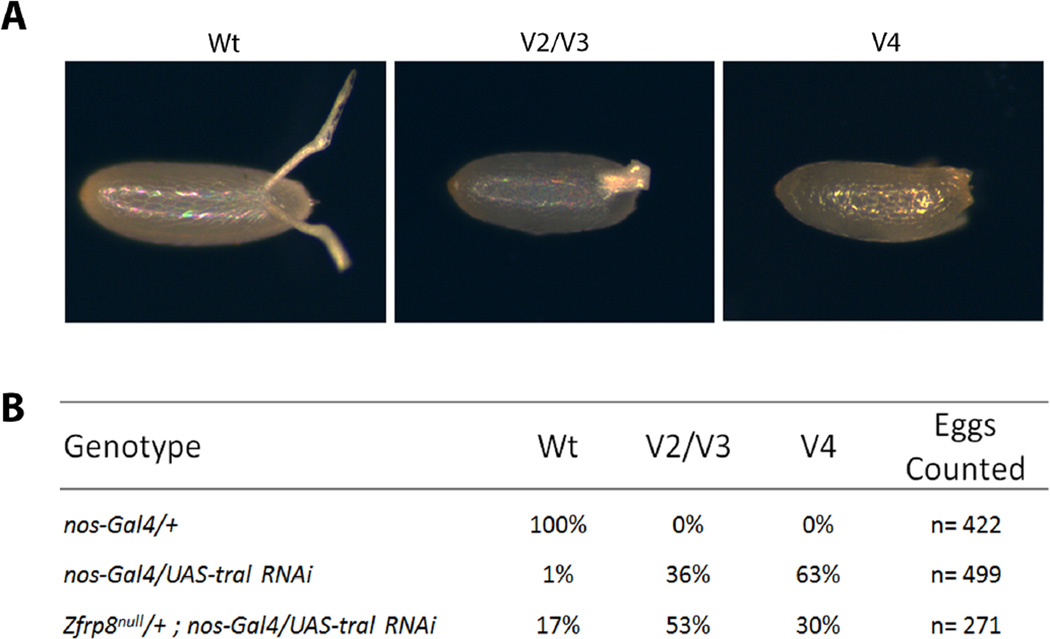
To determine whether Zfrp8 functions in Tral/FMRP-associated translational regulation, we investigated the genetic interaction between Zfrp8 and tral. Tral regulates dorsal-ventral (D/V) patterning through the localization and translational control of gurken (grk) mRNA (Wilhelm et al., 2005). Eggs laid by tral females display ventralized chorion phenotypes, due to the aberrant Gurken morphogen gradient. If Zfrp8 functions to regulate the translational activity of FMRP/Tral, a suppression of the tral ventralized phenotypes should be apparent when Zfrp8 is reduced. We depleted Tral in the germline by expressing a TRiP RNAi line (see Material and Methods) under the control of the nos-Gal4 driver. Tral KD resulted in similar ventralized egg phenotypes as previously observed in eggs laid by tral1 females (Wilhelm et al., 2005): 1% of eggs displayed two normal dorsal appendages (Wt), 36% had fused appendages (category V2/V3), and 63% had no dorsal appendages (category V4, see Materials and Methods) (Fig. 4A–B). Removing one copy of Zfrp8 in the tral KD background suppressed the tral phenotypes (17% Wt, 53% V2/V3, and 30% V4, Fig. 4A–B). This genetic interaction suggests that in addition to controlling the localization of FMRP in the cytoplasm, Zfrp8 also influences the translational control by Tral, essential for formation of dorsal-ventral polarity in the egg (Wilhelm et al., 2005).
Figure 4. Zfrp8 suppresses tral oogenesis phenotypes.
A. w118 (Wt) egg displaying two normally spaced dorsal appendages. A ventralized egg displaying only one wider, fused dorsal appendage (V2/V3) (Li et al., 2014). A fully ventralized egg displaying no dorsal appendages (V4). B. Loss of one copy of Zfrp8 suppresses the tral dorsal-ventral egg phenotype.
We investigated whether Zfrp8 regulates Tral localization as it does FMRP by examining the distribution of GFP-fusion Tral protein trap line (Morin et al., 2001). Tral protein was uniformly present in cytoplasmic compartments of germline and somatic cells, with stronger granules surrounding nuclei, and was highly enriched within the oocyte (Supplemental Fig.3A and 3C). Zfrp8 KD results in loss of oocyte identity (Minakhina et al., 2014), and the distribution of Tral was significantly altered in those cells. But in all other germline cells Tral distribution remained unaffected (Supplemental Fig.3B and 3D). Tral and its orthologs are cytoplasmic proteins (Wilhelm et al., 2005) and examination of the Tral protein sequence identifies no NLSs. Zfrp8 may therefore interact only indirectly with Tral and not regulate its localization.
Zfrp8 and Fmr1 control Position Effect Variegation
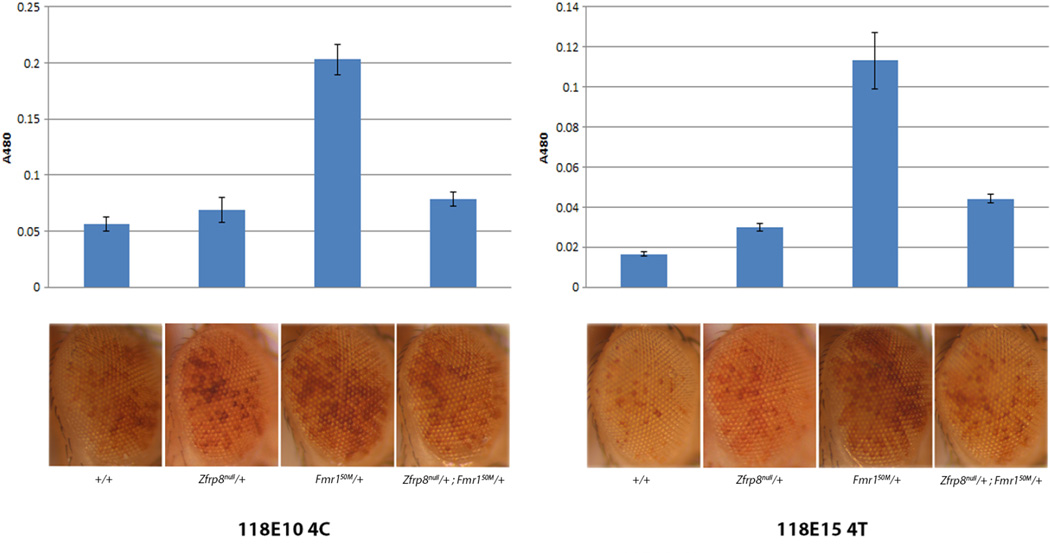
piRNA pathway genes have been shown to be essential for heterochromatin packaging in position effect variegation (PEV) experiments (Brower-Toland et al., 2007; Gu and Elgin, 2013). PEV measures expression of endogenous or reporter genes inserted within or adjacent to heterochromatin. Fmr1 is specifically required for chromatin packaging as loss of a single copy of Fmr1 is sufficient to inhibit heterochromatin silencing of a white reporter (w+) inserted into the pericentric heterochromatin region 118E10 on the 4th chromosome (Deshpande et al., 2006).
We analyzed PEV of Zfrp8 heterozygotes, Fmr1 heterozygotes and Fmr1, Zfrp8 transheterozygotes using 118E10 (4th chromosome centromeric) and an additional w+ reporter, inserted into heterochromatin region 118E15 (4th chromosome telomeric). While the w+ reporters in Zfrp8null/+ eyes were expressed at levels comparable to those in wild-type controls, expression in Fmr1Δ50M /+ of both w+ reporters was strongly enhanced. But, the removal of one copy of both Zfrp8 and Fmr1 decreased expression of the reporters back to the Zfrp8/+, near wild-type levels, indicating restored heterochromatin silencing of both 4th chromosomal insertions (Fig. 5). These findings suggest that in normal eyes, Zfrp8 functions upstream of Fmr1 and controls Fmr1 effects on heterochromatin packaging.
Figure 5. Zfrp8 and Fmr1 control Position Effect Variegation (PEV).
A. Eyes of fly expressing the PEV reporter [w+]118E15-4T in w; w, Zfrp8null/+; w, Fmr150M/+; w, Zfrp8null/+, Fmr150M/+ backgrounds. B. Eyes of fly expressing the PEV reporter [w+]118E10-4C in w; w, Zfrp8null/+; w, Fmr150M/+; w, Zfrp8null/+, Fmr150M/+ backgrounds. The expression of both reporters is unchanged in the Zfrp8null/+ background, while the expression is enhanced in the Fmr150M/+ background. In a Zfrp8null/+, Fmr150M/+ background, expression is suppressed compared to Fmr150M/+ alone. Measurement of eye color (OD 480nm) of extracts from corresponding fly heads support the result.
A connection between regulation of heterochromatin silencing and Piwi has clearly been established and our results show that Zfrp8 and FMRP are part of the mechanism that controls heterochromatin silencing (Brower-Toland et al., 2007; Gu and Elgin, 2013). Heterochromatin is established at the blastoderm stage in Drosophila embryos and is subsequently maintained throughout development. Thus, FMRP and Zfrp8 function together in heterochromatin packaging in the early embryo in the same way as they do during oogenesis.
Conclusion
Here we show that Zfrp8 is part of a complex that is involved in RNA processing, i.e. translation, localization, and stability. We propose that Zfrp8 likely forms a ribonucleoprotein complex with Nufip, FMRP and select mRNAs in the nucleus, and is required for localization of this complex in the cytoplasm. After nuclear export, mRNAs within the complex are targeted for translational control and repression by FMRP and Tral. The suppression of the Fmr1 and tral phenotypes in a Zfrp8 heterozygous background, occurs in the absence of Fmr1 and the strong reduction of tral. This suggests that Zfrp8 function is not protein specific, but rather that it controls the FMRP and Tral-associated complex, even in the absence of each of the two proteins. Our hypothesis is consistent with Zfrp8 actively controlling the localization of FMRP to cytoplasmic foci, as this localization is affected in Zfrp8 germ cells.
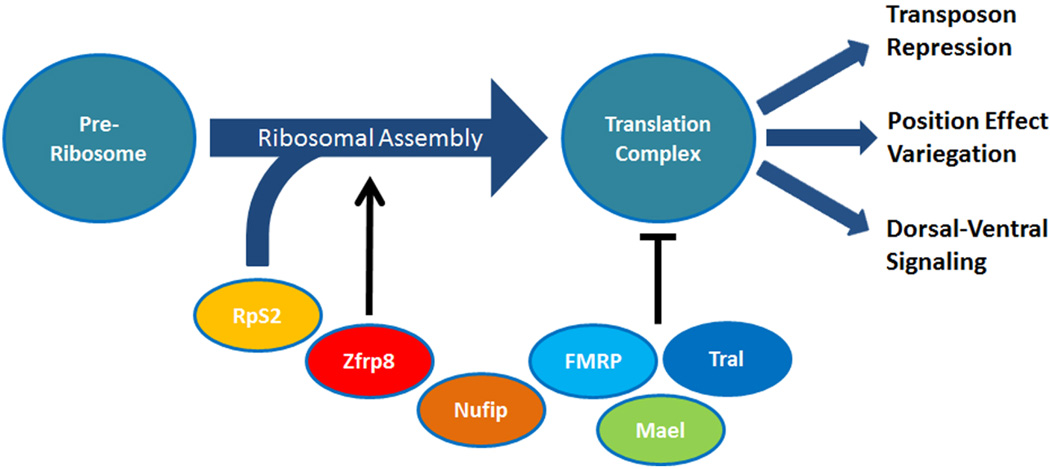
We have previously identified a piRNA pathway protein, Maelstrom (Mael), that is controlled by Zfrp8 in a similar manner as FMRP. Zfrp8 forms a protein complex with Mael, genetically suppresses the loss of mael, and controls Mael localization to the nuage, a perinuclear structure (Minakhina et al., 2014). But the Zfrp8 phenotype is stronger and appears earlier than that of mael, tral, Fmr1, or other piRNA pathway regulatory genes we have studied so far. Zfrp8 may therefore control a central step in the regulation of specific RNPs. Consistent with this hypothesis, our TAP purification and mass spectrometry analysis identified a number of Zfrp8-associated proteins, the majority of which function in ribosomal assembly or translational regulation, such as the ribosomal protein RpS2. And Zfrp8 KD in the germ line and partial loss of rps2 result in a similar “string of pearls phenotype”, caused by developmental arrest in early stages of oogenesis (Cramton and Laski, 1994; Minakhina et al., 2014). In addition, a recent study has shown that Zfrp8 and PDCD2 contain a TYPP (TSR4 in yeast, YwqG in E. coli, PDCD2 and PDCD2L in vertebrates and flies) domain, which has been suggested to perform a chaperone-like function in facilitating protein-protein interactions during RNA processing (Burroughs and Aravind, 2014). These observations lead us to hypothesize that Zfrp8 functions as a chaperone essential for the assembly of ribosomes and the early recruitment and localization of ribosomal-associated regulatory proteins, such as FMRP, Tral and Mael (Fig. 6).
Figure 6. Model of the requirement of Zfrp8 and associated proteins.
Zfrp8 negatively controls the functions of Fmr1 and tral. In the absence of FMRP and Tral the temporal and spatial control of translation of their associated RNPs is lost. We propose that reducing the level of Zfrp8 diminishes the availability of these RNP-complexes in the cytoplasm resulting in suppression of the Fmr1 and tral phenotypes.
Zfrp8, Fmr1 and tral have all been shown to genetically and physically interact with components of the piRNA pathway, and to regulate the expression levels of select transposable elements (Liu et al., 2011; Megosh et al., 2006; Minakhina et al., 2014). Transposon de-repression is often associated with the loss of heterochromatin silencing. The molecular mechanisms underlying heterochromatin formation appear to involve maternally contributed piRNAs and piRNA pathway proteins that control the setting of epigenetic marks in the form of histone modifications, maintained throughout development (Gu and Elgin, 2013). But transposon expression can also be controlled post-transcriptionally by cytoplasmic PIWI-piRNA complexes, suggesting that transposon deregulation and heterochromatin silencing phenotypes seen in FMRP and Zfrp8 may be linked to translational de-repression (Lim et al., 2009; Rouget et al., 2010). We propose that by facilitating the early assembly of ribosomes with specific translational repressors, Zfrp8 regulates several developmental processes during oogenesis and early embryogenesis including dorsal-ventral signaling, transposon de-repression, and position effect variegation.
Supplementary Material
Acknowledgments
We thank Kenneth Irvine, Cordelia Rauskolb, Sarah Radford, Arunika Das, and Fei Wang for helpful comments on the manuscript; Trudi Schupbach, Thomas Jongens, Daniela Zarnescu, Haruhiko Siomi, Lynn Cooley and Phillip Sharp for fly stocks, vectors and antibodies; Le Nguyen for help with stocks and fly food. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We thank Peter Lobel and the Rutgers/RWJMS Biological Mass Spectrometry Facility (supported by a shared instrumentation grant from NIH, S10RR024584) for analysis of the Zfrp8 protein complex. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks used in this study. This work was supported by a grant from the National Institutes of Health (2RO1 GM089992).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, Brand AH, Newbury SF, Wilhelm JE, Levine RB, Nakamura A, Parker R, Ramaswami M. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza N, Minakhina S, Medina DJ, Balsara B, Greenwood S, Huzzy L, Rabson AB, Steward R, Schaar DG. PDCD2 functions in cancer cell proliferation and predicts relapsed leukemia. Cancer Biol Ther. 2013;14:546–555. doi: 10.4161/cbt.24484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoni B, Willemsen R, Weiler IJ, Schenck A, Severijnen LA, Hindelang C, Lalli E, Mandel JL. NUFIP1 (nuclear FMRP interacting protein 1) is a nucleocytoplasmic shuttling protein associated with active synaptoneurosomes. Exp Cell Res. 2003;289:95–107. doi: 10.1016/s0014-4827(03)00222-2. [DOI] [PubMed] [Google Scholar]
- Bauer CR, Epstein AM, Sweeney SJ, Zarnescu DC, Bosco G. Genetic and systems level analysis of Drosophila sticky/citron kinase and dFmr1 mutants reveals common regulation of genetic networks. BMC systems biology. 2008;2:101. doi: 10.1186/1752-0509-2-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Bjorklund M, Furger E, Schertel C, Taipale J, Basler K. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 2013;140:2434–2442. doi: 10.1242/dev.088757. [DOI] [PubMed] [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, Bardoni B, Krol A, Branlant C, Allmang C, Bertrand E, Charpentier B. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. The Journal of cell biology. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzetti MP, Specchia V, Cattenoz PB, Laneve P, Geusa A, Sahin HB, Di Tommaso S, Friscini A, Massari S, Diebold C, Giangrande A. The Drosophila fragile X mental retardation protein participates in the piRNA pathway. Journal of cell science. 2015;128:2070–2084. doi: 10.1242/jcs.161810. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes & development. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Aravind L. Analysis of two domains with novel RNA-processing activities throws light on the complex evolution of ribosomal RNA biogenesis. Front Genet. 2014;5:424. doi: 10.3389/fgene.2014.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan MA, Cabernard C, Heck J, Luois S, Doe CQ, Zarnescu DC. Fragile X protein controls neural stem cell proliferation in the Drosophila brain. Human molecular genetics. 2010;19:3068–3079. doi: 10.1093/hmg/ddq213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Molecular cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yun SW, Seto J, Liu W, Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–1017. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- Costa A, Wang Y, Dockendorff TC, Erdjument-Bromage H, Tempst P, Schedl P, Jongens TA. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Developmental cell. 2005;8:331–342. doi: 10.1016/j.devcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & development. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramton SE, Laski FA. string of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics. 1994;137:1039–1048. doi: 10.1093/genetics/137.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Schedl P. The drosophila fragile X protein dFMR1 is required during early embryogenesis for pole cell formation and rapid nuclear division cycles. Genetics. 2006;174:1287–1298. doi: 10.1534/genetics.106.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Granier CJ, Wang W, Tsang T, Steward R, Sabaawy HE, Bhaumik M, Rabson AB. Conditional inactivation of PDCD2 induces p53 activation and cell cycle arrest. Biology open. 2014;3:821–831. doi: 10.1242/bio.20148326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Elgin SC. Maternal depletion of Piwi, a component of the RNAi system, impacts heterochromatin formation in Drosophila. PLoS genetics. 2013;9:e1003780. doi: 10.1371/journal.pgen.1003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman PJ. The fragile X prevalence paradox. Journal of medical genetics. 2008;45:498–499. doi: 10.1136/jmg.2008.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson AM, Cooley L. Drosophila Kelch functions with Cullin-3 to organize the ring canal actin cytoskeleton. The Journal of cell biology. 2010;188:29–37. doi: 10.1083/jcb.200909017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske M, Moritz B, Anders A, Wahle E. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. The EMBO journal. 2011;30:90–103. doi: 10.1038/emboj.2010.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kidd SA, Lachiewicz A, Barbouth D, Blitz RK, Delahunty C, McBrien D, Visootsak J, Berry-Kravis E. Fragile X syndrome: a review of associated medical problems. Pediatrics. 2014;134:995–1005. doi: 10.1542/peds.2013-4301. [DOI] [PubMed] [Google Scholar]
- Kim M, Bellini M, Ceman S. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Molecular and cellular biology. 2009;29:214–228. doi: 10.1128/MCB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic acids research. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- Kugler JM, Chicoine J, Lasko P. Bicaudal-C associates with a Trailer Hitch/Me31B complex and is required for efficient Gurken secretion. Developmental biology. 2009;328:160–172. doi: 10.1016/j.ydbio.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakakis P, Tipping M, Abed L, Veraksa A. Tandem affinity purification in Drosophila: the advantages of the GS-TAP system. Fly (Austin) 2008;2:229–235. doi: 10.4161/fly.6669. [DOI] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein engineering, design & selection : PEDS. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes & development. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Li W, Klovstad M, Schupbach T. Repression of Gurken translation by a meiotic checkpoint in Drosophila oogenesis is suppressed by a reduction in the dose of eIF1A. Development. 2014;141:3910–3921. doi: 10.1242/dev.109306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. The Journal of cell biology. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development. 2011;138:1863–1873. doi: 10.1242/dev.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Shan G, Guo W, Smrt RD, Johnson EB, Li X, Pfeiffer RL, Szulwach KE, Duan R, Barkho BZ, Li W, Liu C, Jin P, Zhao X. Fragile×mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS genetics. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, Leevers SJ, Cook KR. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Watkins NJ. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Molecular and cellular biology. 2009;29:4971–4981. doi: 10.1128/MCB.00752-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Current biology : CB. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Minakhina S, Changela N, Steward R. Zfrp8/PDCD2 is required in ovarian stem cells and interacts with the piRNA pathway machinery. Development. 2014;141:259–268. doi: 10.1242/dev.101410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakhina S, Steward R. Hematopoietic stem cells in Drosophila. Development. 2010;137:27–31. doi: 10.1242/dev.043943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu W, Munroe RJ, Barker AK, Schimenti JC. PDCD2 is essential for inner cell mass development and embryonic stem cell maintenance. Developmental biology. 2010;347:279–288. doi: 10.1016/j.ydbio.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Pimental W, Tiossi R. Comparison between visual and instrumental methods for natural tooth shade matching. Gen Dent. 2014;62:47–49. [PubMed] [Google Scholar]
- Rothe B, Back R, Quinternet M, Bizarro J, Robert MC, Blaud M, Romier C, Manival X, Charpentier B, Bertrand E, Branlant C. Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly. Nucleic acids research. 2014;42:2015–2036. doi: 10.1093/nar/gkt1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AR, Kanellopoulos AK, Bagni C. Learning and behavioral deficits associated with the absence of the fragile X mental retardation protein: what a fly and mouse model can teach us. Learning & memory. 2014;21:543–555. doi: 10.1101/lm.035956.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr RB, Sharp PA. PDCD2 is a negative regulator of HCF-1 (C1) Oncogene. 2002;21:5245–5254. doi: 10.1038/sj.onc.1205647. [DOI] [PubMed] [Google Scholar]
- Sherman SL, Curnow EC, Easley CA, Jin P, Hukema RK, Tejada MI, Willemsen R, Usdin K. Use of model systems to understand the etiology of fragile X-associated primary ovarian insufficiency (FXPOI) Journal of neurodevelopmental disorders. 2014;6:26. doi: 10.1186/1866-1955-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM. Bicaudal C and trailer hitch have similar roles in gurken mRNA localization and cytoskeletal organization. Developmental biology. 2009;328:434–444. doi: 10.1016/j.ydbio.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha MS, Nouri K, Milroy LG, Moll JM, Herrmann C, Brunsveld L, Piekorz RP, Ahmadian MR. Subcellular fractionation and localization studies reveal a direct interaction of the fragile X mental retardation protein (FMRP) with nucleolin. PLoS One. 2014;9:e91465. doi: 10.1371/journal.pone.0091465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervonen K, Waissi G, Petersen EJ, Akkanen J, Kukkonen JV. Analysis of fullerene-C60 and kinetic measurements for its accumulation and depuration in Daphnia magna. Environ Toxicol Chem. 2010;29:1072–1078. doi: 10.1002/etc.124. [DOI] [PubMed] [Google Scholar]
- Veraksa A, Bauer A, Artavanis-Tsakonas S. Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;232:827–834. doi: 10.1002/dvdy.20272. [DOI] [PubMed] [Google Scholar]
- Wang H, Dictenberg JB, Ku L, Li W, Bassell GJ, Feng Y. Dynamic association of the fragile X mental retardation protein as a messenger ribonucleoprotein between microtubules and polyribosomes. Mol Biol Cell. 2008;19:105–114. doi: 10.1091/mbc.E07-06-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Buszczak M, Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Developmental cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Yang L, Duan R, Chen D, Wang J, Chen D, Jin P. Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila. Human molecular genetics. 2007;16:1814–1820. doi: 10.1093/hmg/ddm129. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Matthies HJ, Mancuso J, Andrews HK, Woodruff E, 3rd, Friedman D, Broadie K. The Drosophila fragile X-related gene regulates axoneme differentiation during spermatogenesis. Developmental biology. 2004;270:290–307. doi: 10.1016/j.ydbio.2004.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.