Abstract
Purpose of Review
Endogenous synthesis of the long chain polyunsaturated fatty acids (LCPUFA) is mediated by the fatty acid desaturase (FADS) gene cluster (11q12-13.1) and elongation of very long chain fatty acids 2 (ELOVL2) (6p24.2) and ELOVL5 (6p12.1). Though older biochemical work identified the product of one gene, FADS2, rate limiting for LCPUFA synthesis, recent studies suggest that polymorphisms in any of these genes can limit accumulation of product LCPUFA.
Recent findings
Genome-wide association study (GWAS) of Greenland Inuit show strong adaptation signals within FADS gene cluster, attributed to high omega-3 fatty acid intake, while GWAS found ELOVL2 associated with sleep duration, age and DNA methylation. ELOVL5 coding mutations cause spinocerebellar ataxia 38, and epigenetic marks were associated with depression and suicide risk. Two sterol response element binding sites were found on ELOVL5, a SREBP-1c target gene. Minor allele carriers of a 3 single nucleotide polymorphism (SNP) haplotype in ELOVL2 have decreased 22:6n-3 levels. Unequivocal molecular evidence shows mammalian FADS2 catalyzes direct Δ4-desaturation to yield 22:6n-3 and 22:5n-6. A SNP near FADS1 influences the levels of 5-lipoxygenase products and epigenetic alteration.
Summary
Genetic polymorphisms within FADS and ELOVL can limit LCPUFA product accumulation at any step of the biosynthetic pathway.
Keywords: desaturase, elongase, long chain polyunsaturated fatty acids (LCPUFA), single nucleotide polymorphisms (SNPs)
INTRODUCTION
Omega3 (ω3 or n-3) and omega6 (ω6 or n-6) long chain polyunsaturated fatty acids (LCPUFA) are ubiquitous in mammalian tissue. They are key nutrients critical for growth and development, are bioactive cellular components of membrane phospholipids, serve as substrates for signaling molecules and act as direct modulators of gene expression [1, 2]. The degree of unsaturation of the biological membranes is modulated by the action of the desaturation and elongation enzymes mediating fatty acid biosynthesis and metabolism. In most organisms endogenous synthesis of LCPUFA from PUFA precursors is possible, but the transformations and efficiencies are specific to cell types and species. In humans, genetic variants within genes encoding for desaturation and elongation enzymes were shown to be associated with LCPUFA levels and complex disease phenotypes. Here, we present recent information gained from studies related to desaturases and elongases limiting endogenous LCPUFA synthesis.
LCPUFA BIOSYNTHESIS
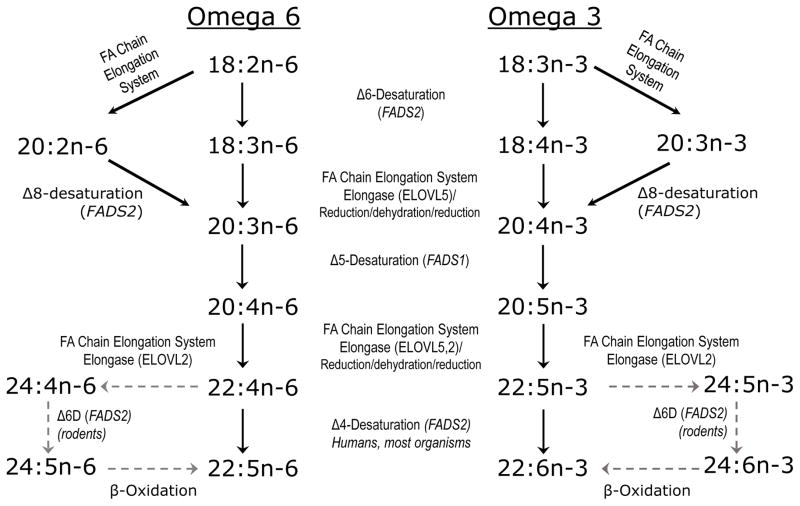
LCPUFA are endogenously biosynthesized from 18:3n-3 and 18:2n-6 PUFA precursors by position-specific desaturation and carbon chain-elongation reactions, as shown in Figure 1. The two PUFA series n-3 and n-6 compete for the same enzymes in the LCPUFA biosynthetic pathway, originally worked out in rodents based on tissue composition resulting from diets rich in 18:3n-3 (alpha-linolenic acid) or 18:2n-6 (linoleic acid). The Δ6-desaturase (fatty acid desaturase 2, FADS2) metabolizes both 18:3n-3 and 18:2n-6, resulting in the synthesis of 6,9,12,15-18:4 and 6,9,12-18:3, respectively. This initial Δ6-desaturation step is widely regarded as rate limiting for LCPUFA endogenous biosynthesis based on biochemical studies, however recent data indicate that other steps in the pathway can limit LCPUFA levels and complex phenotypes. Both 20:5n-3 (eicosapentaenoic acid, EPA) and 20:4n-6 (arachidonic acid) can be further elongated and desaturated to yield 22:6n-3 (docosahexaenoic acid, DHA) and 22:5n-6, respectively. The final steps were long thought to be by direct Δ4-desaturation via 22:5n-3→22:6n-3. Biochemical data in rat liver developed in the 1990s established an alternative coupled microsomal-peroxisomal pathway via 22:5n-3→24:5n-3→24:6n-3→22:6n-3, where the last step is one round of β-oxidation in the peroxisomes [3]. Molecular studies since 2001 established that Δ4-desaturase (FADS2) is the final step in marine microorganisms (e.g.Thraustochytrium), marine teleost fish, and mammals, and very recently in humans [3].
Figure 1.
LCPUFA Biosynthesis Pathway. The omega 6 (n-6) and omega 3 (n-3) fatty acids are substrates in competition for the same sets of FADS and ELOVL catalyzing desaturation and elongation, respectively. Elongation is mediated by a four enzyme coupled system; the first, rate limiting enzyme is the “elongase”.
DESATURASES
Desaturase enzymes perform dehydrogenation reactions and introduce a stereospecific double bond between defined carbons of fatty acyl chains. They have evolved independently twice; the Acyl-acyl carrier protein (ACP) desaturases are soluble enzymes found in the plant plastid and most wide-spread membrane-bound desaturase enzymes found in prokaryotes and eukaryotes [4]. In humans, membrane-bound PUFA desaturases known as “front-end” desaturases introduce a nascent double bond between an existing double bond usually located between the carboxyl group and the 9th carbon atoms from the terminal methyl (n-9) [5]. Front-end desaturation proceeds at the Δ4, Δ5, Δ6 and Δ8 positions and is responsible for endogenous biosynthesis of LCPUFA [3, 6, 7].
FADS1 (Δ5-desaturase), FADS2 (Δ6-desaturase/Δ8-desaturase/Δ4-desaturase) and FADS3 are located as a cluster within 100 kb region on the long arm of human chromosome 11 (HSA11q12-13.1), whereas, mouse Fads homologs with similar structural organization are localized to chromosome 19 [8, 9]. All three genes have evolved by gene duplication events, share 12 exons and 11 introns, and contain well conserved cytochrome b5 domain and three histidine repeats (HXXXH, HXXHH and QXXHH).
FADS2 (Δ6, Δ8, Δ4-DESATURASE)
FADS2 (OMIM#606149) in humans spans 39.1 kb of genomic DNA, encoding a 444-amino acid protein with a molecular mass of 52.3 kDa [8]. FADS2 is a trifunctional even carbon numbered desaturase and acts on at least eleven known fatty acid substrates. In addition to humans, Fads2 has been cloned from mouse, rat and Caenorhabditis elegans [10]. It introduces a double bond at the Δ6 position (between carbons 6 and 7) by acting on at least six substrates (18:2n-6 → 18:3n-6, 18:3n-3 → 18:4n-3, 24:5n-3 → 24:6n-3, 24:4n-6 → 24:5n-6, 16:0 →16:1n-10, and 18:1n-9 →18:2n-9), Δ8-desaturation by acting on 20:1n-9 → 20:2n-9, 20:2n-6 → 20:3n-6, and 20:3n-3 → 20:4n-3, and Δ4-desaturation by acting on 22:4n-6 → 22:5n-6 and 22:5n-3 → 22:6n-3 [3, 10, 11]. The Δ8-desaturase activity provides an alternative pathway to LCPUFA biosynthesis, possibly available when Δ6-desaturase activity is compromised. FADS2 Δ8-desaturates 20:2n-6 and 20:3n-3 to eicosanoid precursors as well as to precursors of 20:4n-6 and 20:5n-3 respectively [10]. This result offers an explanation as to why 20:2n-6 has been associated with FADS single nucleotide polymorphisms (SNPs) in genetic studies described below.
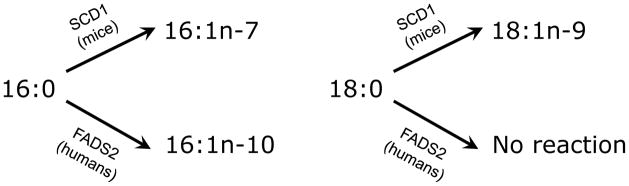
The desaturation of saturated fatty acids (SFA) and PUFA is nearly always considered separately in mammals, with the FADS acting on PUFA and the stearoyl CoA desaturase 1 (SCD1) on SFA. However, 16:0 is the only exception, being a substrate for both Scd1 (16:0→16:1n-7) and FADS2 (16:0→16:1n-10). Human skin lipids, sebum, have long been known as unique with about 25% of fatty acids as 16:1n-10 [12]. Importantly, human and mouse skin cells handle this metabolite shift with different genes: humans use FADS2 to make 16:1n-10 and mice use Scd1 to make 16:1n-7 (Figure 2). Scd1 principally mediates desaturation of 18:0→18:1n-9 (Figure 2), however we detect no activity of FADS2 towards 18:0 in a human cell system.
Figure 2.
Biosynthesis of monounsaturated fatty acids (MUFA) in mouse and human skin. Human skin expresses only FADS2 while mice and all other animals’ skin expresses only Scd1. Both SCD1 and FADS2 are expressed and their resulting enzymes are active in liver and other organs of both humans and mice. Mouse: Scd1 mediates 18:0 conversion to 18:1n-9 and 16:0 conversion to 16:1n-7; Human: FADS2 mediates conversion of 16:0 to 16:1n-10 but has no effect on any other saturated fatty acid (e.g. 18:0 no enzymatic activity).
A human MCF-7 cell with no detectable Δ6-desaturase (FADS2) activity stably transformed with FADS2 mediates direct Δ4 desaturation to yield 22:6n-3 and 22:5n-6, similar to fish and many other organisms [3]. Fads2 null mice had severe problems with fertility; once born, both female and male mice had normal viability and lifespan but were sterile [13]. The Fads2 disruption caused an upstream deficiency in eicosanoid synthesis via reduction in 20:4n-6 substrate, unusual fatty acid biosynthesis, dermal and intestinal ulceration, reduced insulin sensitivity and perturbed cell membrane structure [14, 15]. FADS2 is alternatively spliced to generate two isoforms (FADS2AT1 and FADS2AT2, “AT”=alternative transcript) [16, 17]. We have shown that polypyrimidine tract binding protein (PTB, also known as PTBP1 or hnRNP I) regulates alternative splicing of Fads2. Knock-down of PTB modulated the balance of omega-3 to omega-6 fatty acids by dramatically reducing (50% reduction) 20:5n-3 content [1].
FADS1 (Δ5-DESATURASE)
FADS1 (OMIM#606148) in humans spans 17.2 kb of genomic DNA, encodes a 444-amino acid protein with a molecular mass of 52.0 kDa and shares 61% and 52% identity with FADS2 and FADS3, respectively [8]. It introduces a double bond at the Δ5 position (between carbons 5-6) by acting on at least four 20-carbon fatty acid substrates 20:3n-3, 20:4n-3, 20:2n-6 and 20:3n-6 [6]. When FADS2 is absent, FADS1 produces rare butylene-interrupted fatty acids, for instance 5,11,14-20:3 and 5,11,14,17-20:4. These and similar PUFA are observed in cell systems [6], knockout mice, and normal domestic cats [18]. In this sense, FADS1 competes successfully with FADS2 when FADS2 expression is negligible. Disruption of the Fads1 gene in mouse causes massive accumulation of the 20:3n-6 substrate and 1-series-derived prostaglandins, with a concurrent decrease in the product 20:4n-6 and 2-series-derived prostaglandins [19]. Fads1 ablated mice fail to thrive beyond 12 weeks of age; the phenotype is rescued by dietary supplementation of 20:4n-6 [19]. We recently showed FADS1 producing several mRNA and protein isoforms. One FADS1 isoform (FADS1AT1) enhances desaturation activity of FADS2, leading to increased production of eicosanoid precursors [7].
FADS3
FADS3 (OMIM#606150) is the enigmatic third member of the FADS gene cluster. In humans it spans 17.9 kb of genomic DNA, presumed to encode a 445-amino acid protein with a molecular mass of 51.1 kDa [8]. Fads3 [7, 20] is translated, but no reports exist showing FADS3 mediating front-end desaturation analogous to FADS1 and FADS2. It is extensively spliced, generating at least 8 alternative transcripts that are phylogenetically conserved in several mammalian and avian species [9]. Several FADS3 isoforms have been reported using immunoblotting [20], and are phylogenetically conserved.
Some aspects of its regulation are known and provide clues to its function. Gene transcript studies show Fads3 is highly expressed in mouse uterus at the implantation site; in a Fads2 null mouse, Fads3 expression increased by 3-fold; in baboons fed 22:6n-3 and 20:4n-6 Fads3 ATs abundance increases while Fads1 and Fads2 classical transcripts decrease [9, 21]. An in vitro study provided evidence for Fads3 desaturation of trans11-18:1 (the most abundance trans fatty acid in bovine milkfat) to make a conjugated isomer (trans11,cis13-18:2) by back-end desaturation at position Δ13 (between carbons 13 and 14) common in plants, though the final product structure was not positively identified [22]. We generated the first Fads3 null mouse and found no differences in overt phenotypes (survival, fertility, growth rate) between null and wild type, but fatty acid tissue profiles support a role for Fads3 in the synthesis of DHA during perinatal period [23]. Dosing of trans11-18:1 in aged wild-type mice and comparison to Fads3 null mice provided no in vivo evidence for the 11,13 isomer (Zhang et al, 2015, unpublished observations).
ELONGASES
Fatty acid elongation is well known to occur in cytosol, mitochondria and predominantly microsomes. The microsomal fatty acid chain elongation system (FACES) pathway cycles through a four step process (condensation, reduction, dehydration and reduction) using fatty acids of 12 or more carbons from endogenous and exogenous sources, adding two carbons in each cycle [24]. The first, condensation step is rate limiting and is catalyzed by ELOVL family in mammals, comprised of seven members (ELOVL1-ELOVL7). Among the seven elongases, ELOVL1, ELOVL3, ELOVL6 and ELOVL7 prefer SFA and monounsaturated fatty acids (MUFA) as substrates [24]. ELOVL2 and ELOVL5 are PUFA-specific, whereas ELOVL4 (OMIM#605512) prefer SFA and very long chain PUFA (C28-C38) [24]. A highly conserved HXXHH motif is commonly found in all 7 members [25]. Here we focus on the PUFA specific ELOVL2 and ELOVL5.
ELOVL5 (C18- 20 PUFA ELONGASE)
ELOVL5 (OMIM#611805), is expressed in several human tissues, with highest expression detected in Purkinje cells, lung, testis and adrenal gland. It is specific for 18 and 20 carbons PUFA [24–26]. Microsomes from Elovl5 null mice failed to elongate 18:3n-6 to 20:3n-6 and 18:4n-3 to 20:4n-3 resulting in accumulation of 18:3n-6 and 18:4n-3 respectively, and significant lowering of their downstream products 20:4n-6 and 22:6n-3, respectively. These mice develop hepatic steatosis apparently as a consequence of decreased cellular 20:4n-6 and 22:6n-3 and upregulation of sterol regulatory element-binding protein 1c (Srebp-1c) and its target genes [24]. The expression of ELOVL5 is transcriptionally regulated by SREBP-1c and a recent study in human showed existence of two novel sterol regulatory element (SRE) binding sites, one in the upstream region and one in the exon 1 of ELOVL5 [25].
ELOVL2 (C20-24 PUFA ELONGASE)
ELOVL2 (OMIM#611814), is selectively expressed in human tissues, with highest expression detected in testis and liver. It has substrate specificities for 20 and 22 carbon PUFA [24, 27–29]. Rat Elovl2 converts 22:5n-3 to 24:5n-3 [28], whereas, in chickens both Elovl2 and Elovl5 convert 22:5n-3 to 24:5n-3 demonstrating species specific differences [30]. Ablation of Elovl2 caused complete arrest of spermatogenesis, complete absence of very long chain PUFA (carbon chain length between 24 to 30) of the n-6 family in testis and significant increase in the serum levels of 20:5n-3 and 22:5n-3, with concurrent non-significant decrease in 22:6n-3 [31]. The supplementation of 22:6n-3 for 3 months was not able to restore male fertility in these mice [31]. In the follow-up study the same group found Elovl2 deletion caused severe reduction of 22:6n-3 and 22:5n-6 and an accumulation of 22:5n-3 and 22:4n-6 in both liver and serum. These mice had increased expression of Srebp-1c and its target genes (Fasn and Scd1) in liver but did not develop steatosis [32].
GENETIC VARIANTS: LCPUFA LEVELS AND HUMAN PHENOTYPES
The FADS and ELOVL are among the most prominent genes associated with human phenotypes in both candidate gene study and genome-wide association study (GWAS). It has long been known that carnivores, such as cats and higher trophic level fish, have lost the metabolic ability to synthesize long chain PUFA via loss of Fads2 desaturation activity; presumably this is due to the ubiquitous presence of 20:4n-6 and 22:6n-3 in a meat based diet. In contrast, herbivores ingest very little 20:4n-6 and 22:6n-3 and must have a robust metabolic pathway to synthesize all they need, especially at life stages of high demand such as the brain growth spurt. The remarkable flexibility of humans to survive in environments that predominantly produce animal foods or plant foods suggests adaptive changes specifically in the FADS and ELOVL genes [33].
Converging evidence from candidate gene and GWAS available recently show large genetic variability in the level of fatty acid precursors 18:3n-3 and 18:2n-6 and their apparent conversion to physiologically important LCPUFA products, especially 20:5n-3 and 20:4n-6. These studies have shown strong associations between the minor allele carriers of single nucleotide polymorphisms (SNPs) within FADS gene cluster, ELOVL2 and ELOVL5 and fatty acid levels in serum, plasma, red cells, breast milk and adipose tissue [13, 34–36].
CANDIDATE (FADS GENE CLUSTER, ELOVL2 AND ELOVL5) GENE STUDIES
A FADS gene cluster association study showing minor allele carriers of a 11 SNP haplotype exhibited increased levels of 18:2n-6, 20:2n-6, 20:3n-6, and 18:3n-3 and decreased levels of 18:3n-6, 20:4n-6, and 20:5n-3 in serum [34]. This finding was subsequently replicated independently by others. Genetic variability was highest (28%) for 20:4n-6 [34]. Locus specific (FADS gene cluster) SNPs are associated with human phenotypes, for instance, inflammation and cardiovascular disorders [13, 34], levels of blood lipids (total cholesterol (TC), high-density lipoprotein (HDL), low-density lipoprotein (LCL) and triglyceride(TG)) [37], insulin resistance in schizophrenia and bipolar patients [38], perinatal depression [13], atopic diseases [13, 34], attention/hyperactivity [13] and intelligence in children [13]. A SNP in ELOVL5 was associated with late onset primary open-angle glaucoma [39]. Recent studies show SNPs near FADS1 (rs174537) influencing the levels of 5-lipoxygenase products [40] and another found rs174537 associated with epigenetic alteration [41]. A minor allele of FADS3 SNP (rs174455) was negatively associated with 22:6n-3 in erythrocyte phospholipids in the AVON longitudinal study of parents and children (ALSPAC) cohort [42]. A very distinct FADS haplotype was shown to be associated with enhanced ability to biosynthesize LCPUFA from PUFA precursors [43]. Major allele homozygotes of a 4 SNP haplotype showed more than 3-fold greater apparent conversion of [U-13C]-18:3n-3 to [U-13C]-20:5n-3 in plasma than minor allele carriers [44]. Al Saleh et al. [36] found minor allele carriers of 3 SNP haplotype within the ELOVL2 gene with lower plasma 22:6n-3 than major allele carrier. An SNP within ELOVL5 was nominally associated with decreased capacity to metabolize 20:5n-3 to 22:5n-3 [35]. Epigenetic marks within the regulatory regions of ELOVL5 were associated with depression and suicide risk [45].
GWAS
As a complement to locus specific SNPs, several GWAS identified FADS and ELOVL loci to be associated with LCPUFA levels and human phenotypes. Traditional inhabitants of the arctic region of North America are known to subsist almost exclusively on animal foods, predominantly fish and marine mammals. GWAS show a striking adaptation of the Greenland Inuit FADS gene cluster attributed to high omega-3 fatty acid intake [46]. A meta-analysis of seven GWAS scans and replication with 20,623 individuals identified 30 loci including the FADS cluster as influencing HDL and triglyceride levels [13]. The InCHIANTI GWAS identified a minor allele of rs174537 accounted for 18.6% genetic variance in 20:4n-6 concentrations, which was confirmed in an independent sample from the GOLDN study [13] and another study found genetic variants within FADS1 and FADS2 to be associated with higher levels of ALA and lower levels of EPA and DPA and these associations were similar in ancestries of European, African, Chinese and Hispanic origin [47]. Lemaitre et al.[47] also showed minor alleles of SNPs in ELOVL2 to be associated with lower 22:6n-3 levels, however, these associations were found to be less consistent in the four ancestries. Moreover, there are numerous GWAS reports that relate FADS and ELOVL SNPs to TC-HDL-LDL-TG and lipid metabolite levels [13, 48, 49], fasting glucose homeostasis [50], and resting heart rate [51]. A few studies tried to combine GWAS with metabolomics (mGWAS) to understand gene-environment impacts on homeostasis and to address missing heritability [52]. Genetic information at the FADS1 locus (rs174547) combined with targeted metabolomics identified 36% of the observed variance in metabolite concentrations [53], in the same study SNP (rs9393903) in ELOVL2 accounted for 9.8% variance. Similarly, Suhre et al. [54] showed “genetically determined metabotypes” accounted for 10–60% differences in metabolite levels per allele copy in 25 loci which also includes FADS1 and ELOVL2. FADS1 SNP (rs174548) accounted for 29% of the metabolic ratio variance [13]. ELOVL2 is associated with sleep duration [55], age and DNA methylation [56].
CODING GENETIC VARIANTS (FADS GENE CLUSTER, ELOVL2 AND ELOVL5)
Target captured exome sequencing showed existence of 53,081 coding SNPs (cSNPs) in the human genome [57]. cSNPs may be synonymous, resulting in no change in the resulting protein primary sequence, or non-synonymous, resulting in a substitution of one amino acid for the other at the relevant position. An October 2015 check of the NCBI dbSNP database reveals several cSNPs within the FADS and ELOVL genes. No disease causing phenotypes associated with cSNPs in the FADS gene cluster and ELOVL2 are yet known, however, a FADS2 promoter SNP (rs968567) enhancing FADS2 expression has been discovered [58]. Two non-synonymous mutations within ELOVL5 causing spinocerebellar ataxia 38 (SCA38) has been reported in families originating from Italy and France [26].
INSERTION-DELETION (INDEL) POLYMORPHISM
GWAS and candidate gene studies target SNP(s) that are tags for haplotypes which serve only as signals close to functional variants responsible for phenotypic variation. Association strength is directly related to the physical closeness of the GWAS SNP(s) to the functional element, a basic principle underlying the Manhattan plot.
In humans, highly polymorphic small Indels (1 bp to 10,000 bp) are the second most frequent polymorphisms after SNPs and are increasingly recognized as functional contributors of genetic variation influencing multiple human phenotypes [59]. Largely due to the technical difficulties in genotyping and calling Indels from short read sequencing data, their functional effects are understood only in a few cases [59]. Recently, we discovered a 22-bp Indel polymorphic variant (rs66698963) in FADS2 intron 1 near a SRE to be associated with desaturase expression depending on levels of SREBP-1c agonists [60]. Follow-up work showed the FADS2 Indel strongly influencing metabolic capacity to synthesize 20:4n-6 from precursors and also showed ethnic differences in the allele frequency in general US and Indian subjects, as well as evidence for adaptive evolution (Kothapalli et al., 2015, unpublished data). Commonly reported SNP variants within intron 1 of FADS2 (rs174575, rs174570 and rs1535) and thus the nearby Indel are associated with increased IQ scores, blood fatty acid levels and complex diseases. rs174575 and rs174570 are within 600 and 6000 bp upstream from the FADS2 Indel, respectively [13, 34, 46]. In humans, common SNP variants are often found to follow Indels [61], suggesting that rs174575, rs174570 and/or rs1535 are tags for the functional genomic Indel that directly modulates binding at the nearby SRE. Whether or not the Indel is the functional element or it is nearby, genotypic variation controlling basal concentrations of 20:4n-6, its immediate precursor and its products, demonstrates in vivo that FADS2 protein(s) is (are) a major rate limiting enzyme for LCPUFA production.
CONCLUSION
The genes mediating the endogenous synthesis of LCPUFA contribute wide variability to the efficiency of LCPUFA synthesis, likely controlled at FADS2 but also controlled at the level of FADS1 and the elongases depending on genotype and metabolic state. In the era of mass individual immigration and international food supplies, individuals with genotypes adapted to a food supply with high, or low, LCPUFA will find themselves exposed to diets to which they are not adapted. Precision nutrition depends on the detailed genetics controlling LCPUFA conversion efficiency, for instance, those adapted to high intakes of 20:4n-6 and 22:6n-3 via meat and fish may become deficient when consuming an otherwise heart healthy diet predominantly composed of vegetables; the risk of neurodegenerative diseases may thereby increase. Continuing mechanistic studies of genetic function are needed to address this issue.
KEY POINTS.
Endogenous synthesis of long chain polyunsaturated fatty acids (LCPUFA) and degree of unsaturation of the biological membranes depends largely on the action of the fatty acid desaturases (FADS1, FADS2 and putative FADS3) and elongation of very long chain fatty acids (ELOVL2 and ELOVL5).
While FADS2 mediated Δ6-desaturation is demonstrably rate limiting for LCPUFA synthesis in rodent biochemical studies, emerging human genomic, mouse gene ablation, and in vitro studies indicate that metabolic control over LCPUFA homeostasis is associated with FADS and ELOVL for endogenous synthesis.
Polymorphisms in the FADS gene cluster yield the strongest signal in genomic studies of LCPUFA eating Greenland Inuit people consistent with decades old predictions.
The FADS gene cluster, ELOVL2 and ELOVL5 are associated with LCPUFA levels and complex human phenotypes.
Novel substrate and positional desaturase specificities mediated by FADS2, Δ4 acting on 22:5n-3 and 22:4n-6, and Δ8 acting on 20:2n-6 and 20:3n-3, imply that LCPUFA endogenous synthesis is controlled at several levels.
Acknowledgments
This review was supported by NIH grant R01 AT007003 from the National Center for Complementary and Integrative Health (NCCIH) (formerly the National Center for Complementary and Alternative Medicine (NCCAM)) and the Office of Dietary Supplements (ODS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES AND RECOMMENDED READING
• of special interest
•• of outstanding interest
- 1.Reardon HT, Park WJ, Zhang J, et al. The polypyrimidine tract binding protein regulates desaturase alternative splicing and PUFA composition. J Lipid Res. 2011;52:2279–2286. doi: 10.1194/jlr.M019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qawasmi A, Landeros-Weisenberger A, Leckman JF, Bloch MH. Meta-analysis of long-chain polyunsaturated fatty acid supplementation of formula and infant cognition. Pediatrics. 2012;129:1141–1149. doi: 10.1542/peds.2011-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3••.Park HG, Park WJ, Kothapalli KS, Brenna JT. The fatty acid desaturase 2 (FADS2) gene product catalyzes Delta4 desaturation to yield n-3 docosahexaenoic acid and n-6 docosapentaenoic acid in human cells. FASEB J. 2015;29:3911–3919. doi: 10.1096/fj.15-271783. The FADS2 protein from the classical transcript catalyzes direct Δ4-desaturation of 22:5n-3 to 22:6n-3 (DHA) and 22:4n-6 to 22:5n-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shanklin J, Guy JE, Mishra G, Lindqvist Y. Desaturases: emerging models for understanding functional diversification of diiron-containing enzymes. J Biol Chem. 2009;284:18559–18563. doi: 10.1074/jbc.R900009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meesapyodsuk D, Qiu X. The front-end desaturase: structure, function, evolution and biotechnological use. Lipids. 2012;47:227–237. doi: 10.1007/s11745-011-3617-2. [DOI] [PubMed] [Google Scholar]
- 6.Park WJ, Kothapalli KS, Lawrence P, Brenna JT. FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual eicosanoid fatty acids. PLoS One. 2011;6:e28186. doi: 10.1371/journal.pone.0028186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park WJ, Kothapalli KS, Reardon HT, et al. A novel FADS1 isoform potentiates FADS2-mediated production of eicosanoid precursor fatty acids. J Lipid Res. 2012;53:1502–1512. doi: 10.1194/jlr.M025312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marquardt A, Stohr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 9.Brenna JT, Kothapalli KS, Park WJ. Alternative transcripts of fatty acid desaturase (FADS) genes. Prostaglandins Leukot Essent Fatty Acids. 2010;82:281–285. doi: 10.1016/j.plefa.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park WJ, Kothapalli KS, Lawrence P, et al. An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J Lipid Res. 2009;50:1195–1202. doi: 10.1194/jlr.M800630-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Ichi I, Kono N, Arita Y, et al. Identification of genes and pathways involved in the synthesis of Mead acid (20:3n-9), an indicator of essential fatty acid deficiency. Biochim Biophys Acta. 2014;1841:204–213. doi: 10.1016/j.bbalip.2013.10.013. In vitro siRNA-mediated knock-down experiments indicate two pathways for the biosynthesis of Mead acid are mediated by FADS1, FADS2 and ELOVL5 genes. [DOI] [PubMed] [Google Scholar]
- 12.Park HG, Kothapalli KS, Park WJ, et al. Palmitic acid (16:0) competes with omega-6 linoleic and omega-3 a-linolenic acids for FADS2 mediated Delta6-desaturation. Biochim Biophys Acta. 2015;1861:91–97. doi: 10.1016/j.bbalip.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lattka E, Illig T, Heinrich J, Koletzko B. Do FADS genotypes enhance our knowledge about fatty acid related phenotypes? Clin Nutr. 2010;29:277–287. doi: 10.1016/j.clnu.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Roqueta-Rivera M, Abbott TL, Sivaguru M, et al. Deficiency in the omega-3 fatty acid pathway results in failure of acrosome biogenesis in mice. Biol Reprod. 2011;85:721–732. doi: 10.1095/biolreprod.110.089524. [DOI] [PubMed] [Google Scholar]
- 15•.Stoffel W, Hammels I, Jenke B, et al. Obesity resistance and deregulation of lipogenesis in Delta6-fatty acid desaturase (FADS2) deficiency. EMBO Rep. 2014;15:110–120. doi: 10.1002/embr.201338041. Obesity resistance is a major feature of the Fads2 null mouse phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park WJ, Reardon HT, Tyburczy C, et al. Alternative splicing generates a novel FADS2 alternative transcript in baboons. Mol Biol Rep. 2010;37:2403–2406. doi: 10.1007/s11033-009-9750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kothapalli KS, Guo XX, Sun XX, et al. Alternative transcripts in the human milk fat globule proteinogenic RNA transcriptome with emphasis on desaturases. FASEB J. 2014;28:S818.818. [Google Scholar]
- 18.Trevizan L, de Mello Kessler A, Brenna JT, et al. Maintenance of arachidonic acid and evidence of Delta5 desaturation in cats fed gamma-linolenic and linoleic acid enriched diets. Lipids. 2012;47:413–423. doi: 10.1007/s11745-011-3651-0. [DOI] [PubMed] [Google Scholar]
- 19.Fan YY, Monk JM, Hou TY, et al. Characterization of an arachidonic acid-deficient (Fads1 knockout) mouse model. J Lipid Res. 2012;53:1287–1295. doi: 10.1194/jlr.M024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedrono F, Blanchard H, Kloareg M, et al. The fatty acid desaturase 3 gene encodes for different FADS3 protein isoforms in mammalian tissues. J Lipid Res. 2010;51:472–479. doi: 10.1194/jlr.M000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reardon HT, Hsieh AT, Park WJ, et al. Dietary long-chain polyunsaturated fatty acids upregulate expression of FADS3 transcripts. Prostaglandins Leukot Essent Fatty Acids. 2013;88:15–19. doi: 10.1016/j.plefa.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rioux V, Pedrono F, Blanchard H, et al. Trans-vaccenate is Delta13-desaturated by FADS3 in rodents. J Lipid Res. 2013;54:3438–3452. doi: 10.1194/jlr.M042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JY, Qin X, Liang A, et al. Fatty acid desaturase 3 null mouse biochemical phenotype. FASEB J. 2014;28:S246.245. [Google Scholar]
- 24.Guillou H, Zadravec D, Martin PG, Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 25•.Shikama A, Shinozaki H, Takeuchi Y, et al. Identification of human ELOVL5 enhancer regions controlled by SREBP. Biochem Biophys Res Commun. 2015;465:857–863. doi: 10.1016/j.bbrc.2015.08.101. A detailed promoter/enhancer analysis show the existence of two new sterol response element (SRE) binding sites, one 10 kb upstream and one in the exon1 of ELOVL5, long known as an SREBP-1c target. [DOI] [PubMed] [Google Scholar]
- 26••.Di Gregorio E, Borroni B, Giorgio E, et al. ELOVL5 mutations cause spinocerebellar ataxia 38. Am J Hum Genet. 2014;95:209–217. doi: 10.1016/j.ajhg.2014.07.001. Two coding missense mutations within ELOVL5 gene which are predicted to be damaging were found for the first time in humans of Italian and French ancestry suffering from the spinocerebellar ataxia 38 (SCA38) neurodegenerative disorder. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno Y, Suto S, Yamanaka M, et al. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci U S A. 2010;107:18439–18444. doi: 10.1073/pnas.1005572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory MK, Cleland LG, James MJ. Molecular basis for differential elongation of omega-3 docosapentaenoic acid by the rat Elovl5 and Elovl2. J Lipid Res. 2013;54:2851–2857. doi: 10.1194/jlr.M041368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregory MK, Gibson RA, Cook-Johnson RJ, et al. Elongase reactions as control points in long-chain polyunsaturated fatty acid synthesis. PLoS One. 2011;6:e29662. doi: 10.1371/journal.pone.0029662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregory MK, Geier MS, Gibson RA, James MJ. Functional characterization of the chicken fatty acid elongases. J Nutr. 2013;143:12–16. doi: 10.3945/jn.112.170290. [DOI] [PubMed] [Google Scholar]
- 31.Zadravec D, Tvrdik P, Guillou H, et al. ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. J Lipid Res. 2011;52:245–255. doi: 10.1194/jlr.M011346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Pauter AM, Olsson P, Asadi A, et al. Elovl2 ablation demonstrates that systemic DHA is endogenously produced and is essential for lipid homeostasis in mice. J Lipid Res. 2014;55:718–728. doi: 10.1194/jlr.M046151. Elovl2 ablation causes severe reduction of 22:6n-3 and 22:5n-6 in both liver and serum, with increased expression of SREBP-1c and its liver target genes (Fasn and Scd1); nevertheless mice are resistant to liver steatosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenna JT, Carlson SE. Docosahexaenoic acid and human brain development: evidence that a dietary supply is needed for optimal development. J Hum Evol. 2014;77:99–106. doi: 10.1016/j.jhevol.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Lattka E, Illig T, Koletzko B, Heinrich J. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol. 2010;21:64–69. doi: 10.1097/MOL.0b013e3283327ca8. [DOI] [PubMed] [Google Scholar]
- 35.Morales E, Bustamante M, Gonzalez JR, et al. Genetic variants of the FADS gene cluster and ELOVL gene family, colostrums LC-PUFA levels, breastfeeding, and child cognition. PLoS One. 2011;6:e17181. doi: 10.1371/journal.pone.0017181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Alsaleh A, Maniou Z, Lewis FJ, et al. ELOVL2 gene polymorphisms are associated with increases in plasma eicosapentaenoic and docosahexaenoic acid proportions after fish oil supplement. Genes Nutr. 2014;9:362. doi: 10.1007/s12263-013-0362-6. Minor allele carriers of a 3 SNP ELOVL2 haplotype show lower plasma DHA levels than major allele carrier at baseline. A dose of 1.8 g/day of EPA and DHA causes a greater increase EPA (30%) and DHA (9%) in minor allele carriers compared to non-carriers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Standl M, Lattka E, Stach B, et al. FADS1 FADS2 gene cluster, PUFA intake and blood lipids in children: results from the GINIplus and LISAplus studies. PLoS One. 2012;7:e37780. doi: 10.1371/journal.pone.0037780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burghardt KJ, Gardner KN, Johnson JW, Ellingrod VL. Fatty Acid desaturase gene polymorphisms and metabolic measures in schizophrenia and bipolar patients taking antipsychotics. Cardiovasc Psychiatry Neurol. 2013;2013:596945. doi: 10.1155/2013/596945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mabuchi F, Sakurada Y, Kashiwagi K, et al. Association between SRBD1 and ELOVL5 gene polymorphisms and primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2011;52:4626–4629. doi: 10.1167/iovs.11-7382. [DOI] [PubMed] [Google Scholar]
- 40•.Hester AG, Murphy RC, Uhlson CJ, et al. Relationship between a Common Variant in the Fatty Acid Desaturase (FADS) Cluster and Eicosanoid Generation in Humans. J Biol Chem. 2014;289:22482–22489. doi: 10.1074/jbc.M114.579557. A SNP near FADS1 is associated with the synthesis of arachidonic acid and 5-lipoxygenase products. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Howard TD, Mathias RA, Seeds MC, et al. DNA methylation in an enhancer region of the FADS cluster is associated with FADS activity in human liver. PLoS One. 2014;9:e97510. doi: 10.1371/journal.pone.0097510. Genome-wide allele-specific methylation (ASM) analysis identify rs174537 near FADS1 as methylation-altering SNP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koletzko B, Lattka E, Zeilinger S, et al. Genetic variants of the fatty acid desaturase gene cluster predict amounts of red blood cell docosahexaenoic and other polyunsaturated fatty acids in pregnant women: findings from the Avon Longitudinal Study of Parents and Children. Am J Clin Nutr. 2011;93:211–219. doi: 10.3945/ajcn.110.006189. [DOI] [PubMed] [Google Scholar]
- 43.Ameur A, Enroth S, Johansson A, et al. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet. 2012;90:809–820. doi: 10.1016/j.ajhg.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillingham LG, Harding SV, Rideout TC, et al. Dietary oils and FADS1-FADS2 genetic variants modulate [13C]alpha-linolenic acid metabolism and plasma fatty acid composition. Am J Clin Nutr. 2013;97:195–207. doi: 10.3945/ajcn.112.043117. [DOI] [PubMed] [Google Scholar]
- 45•.Haghighi F, Galfalvy H, Chen S, et al. DNA methylation perturbations in genes involved in polyunsaturated Fatty Acid biosynthesis associated with depression and suicide risk. Front Neurol. 2015;6:92. doi: 10.3389/fneur.2015.00092. PUFA levels and DNA methylation within the regulatory regions of Elovl5 are associated with suicide risk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Fumagalli M, Moltke I, Grarup N, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349:1343–1347. doi: 10.1126/science.aab2319. Genomic analysis of Greenland Inuit with heritage of nearly exclusive carnivory of marine mammals and fish rich in EPA and DHA show strongest adaptive signal in the FADS gene cluster, as predicted by classical studies decades ago. [DOI] [PubMed] [Google Scholar]
- 47.Lemaitre RN, Tanaka T, Tang W, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 2011;7:e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambers JC, Zhang W, Sehmi J, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mirkov S, Myers JL, Ramirez J, Liu W. SNPs affecting serum metabolomic traits may regulate gene transcription and lipid accumulation in the liver. Metabolism. 2012;61:1523–1527. doi: 10.1016/j.metabol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eijgelsheim M, Newton-Cheh C, Sotoodehnia N, et al. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum Mol Genet. 2010;19:3885–3894. doi: 10.1093/hmg/ddq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adamski J. Genome-wide association studies with metabolomics. Genome Med. 2012;4:34. doi: 10.1186/gm333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Illig T, Gieger C, Zhai G, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suhre K, Shin SY, Petersen AK, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheinfeldt LB, Gharani N, Kasper RS, et al. Using the Coriell Personalized Medicine Collaborative Data to conduct a genome-wide association study of sleep duration. Am J Med Genet B Neuropsychiatr Genet 2015. • GWAS using data from participants in the Coriell Personalized Medicine Collaborative (CPMC) identify ELOVL2 as a novel candidate gene for genetic contribution to variation in sleep duration. doi: 10.1002/ajmg.b.32362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Ronn T, Volkov P, Gillberg L, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24:3792–3813. doi: 10.1093/hmg/ddv124. Genome-wide DNA methylation sites analysis in human adipose tissue found ELOVL2 to be the most significantly associated gene with age and DNA methylation. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Vinckenbosch N, Tian G, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 58.Lattka E, Eggers S, Moeller G, et al. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J Lipid Res. 2010;51:182–191. doi: 10.1194/jlr.M900289-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Montgomery SB, Goode DL, Kvikstad E, et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013;23:749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reardon HT, Zhang J, Kothapalli KS, et al. Insertion-deletions in a FADS2 intron 1 conserved regulatory locus control expression of fatty acid desaturases 1 and 2 and modulate response to simvastatin. Prostaglandins Leukot Essent Fatty Acids. 2012;87:25–33. doi: 10.1016/j.plefa.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu JT, Wang Y, Gibbs RA, Yu F. Characterizing linkage disequilibrium and evaluating imputation power of human genomic insertion-deletion polymorphisms. Genome Biol. 2012;13:R15. doi: 10.1186/gb-2012-13-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]