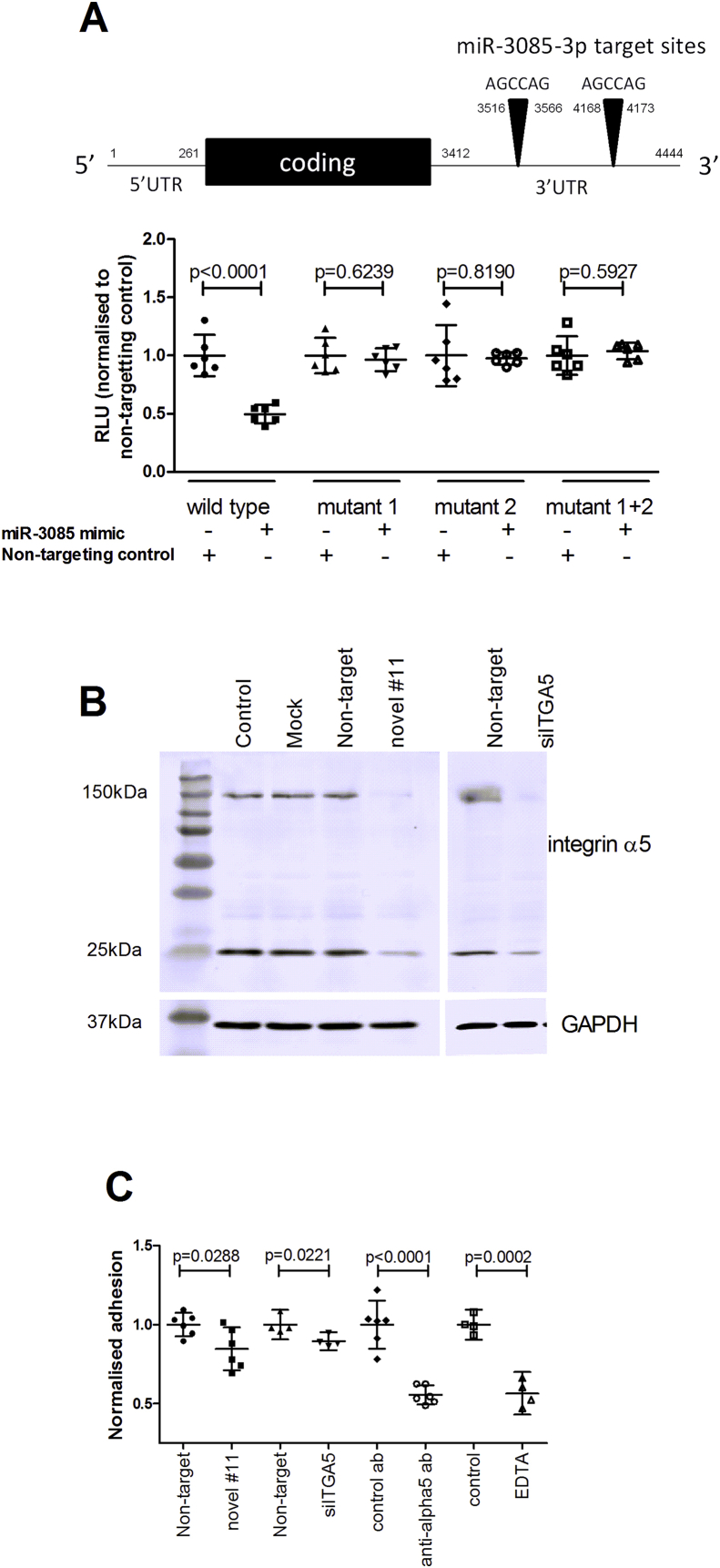
Fig. 5.
Functional analysis of integrin alpha5 as a target of candidate microRNA #11. (A) The schematic shows the ITGA5 cDNA with the location of target sites numbered. Cells (DF1) were transfected with the 3′UTR of ITGA5 cloned into pmiRGLO, or a construct with either or both miR-3085-3p seed sites mutated (to GAGCTC) (mutant 1, 2 or 1 + 2), with or without candidate miRNA #11 (miR-3085-3p) mimic or non-targeting control. Firefly luciferase relative light units were normalised to Renilla relative light units. Data are mean ± 95% confidence interval, n = 6, analysed by a Student's t test. (B) SW1353 cells had no treatment (control), mock transfected, or transfected with a miR-3085 mimic, an siRNA against ITGA5 (siITGA5) or non-targeting siRNA; protein was extracted and separated on 12.5% (w/v) SDS-PAGE, blotted onto PVDF and probed with an anti-integrin alpha5 antibody. The blot was stripped and re-probed with a GAPDH antibody to assess loading. (C) SW1353 cells were transfected with a miR-3085 mimic, siITGA5 or a non-targeting control (100 nM) for 48 h. Cells were pre-incubated in serum-free medium with an anti-integrin alpha5 antibody or control antibody (10 μg/ml) or EDTA (2 mM) for 10 min if required and then allowed to adhere to fibronectin for 15 min. After washing cells were fixed and stained with methylene blue, lysed and absorbance at 590 nm measured. Data are normalised to the control and presented as mean ± 95% confidence interval, n = 4–6, analysed by a Student's t test).