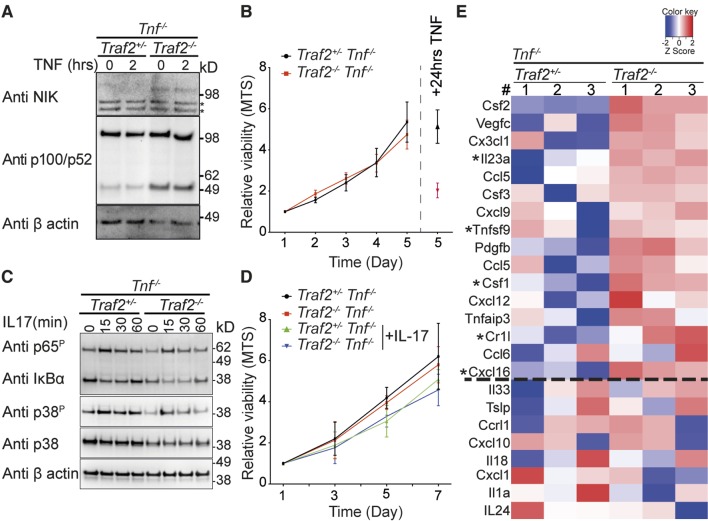
Figure 8. TRAF2 deletion causes constitutive activation of the non-canonical NF-κB transcription factor pathway.
(A) Tnf-/-Traf2+/- and Tnf-/-Traf2-/- keratinocytes were treated with or without TNF (100ng/ml) for the indicated times. The lysates were analysed as in Figure 1. Asterisks indicate non-specific bands. (B) The viability of keratinocytes of the indicated genotype was measured at indicated time points using an MTS assay. Viability following TNF treatment (100 ng/ml) on day 4 was used as a control. Data are represented as mean ± SEM, n=4 biological repeats. (C) Tnf-/-Traf2+/- and Tnf-/-Traf2-/- keratinocytes were treated with IL-17 (100 ng/ml) for the indicated times and lysates analysed as previously. (D) Viability of keratinocytes of indicated genotype ± IL-17 (100 ng/ml) was measured at indicated time points using MTS assay. Data are represented as mean ± SEM, n≥3 biological repeats. (E) Heat map depicting a selection from qPCR analysis of more than 600 inflammatory genes from Tnf-/-Traf2+/- and Tnf-/-Traf2-/- keratinocytes (3 mice for each genotype). Log expression values have been standardized to have mean 0 and standard deviation 1 for each row. Genes were ranked based on the fold change expression. Gens above the dashed line were highly elevated in Tnf-/-Traf2-/- keratinocytes. Asterisks indicate significant changes with P values <0.05.
DOI: http://dx.doi.org/10.7554/eLife.10592.010