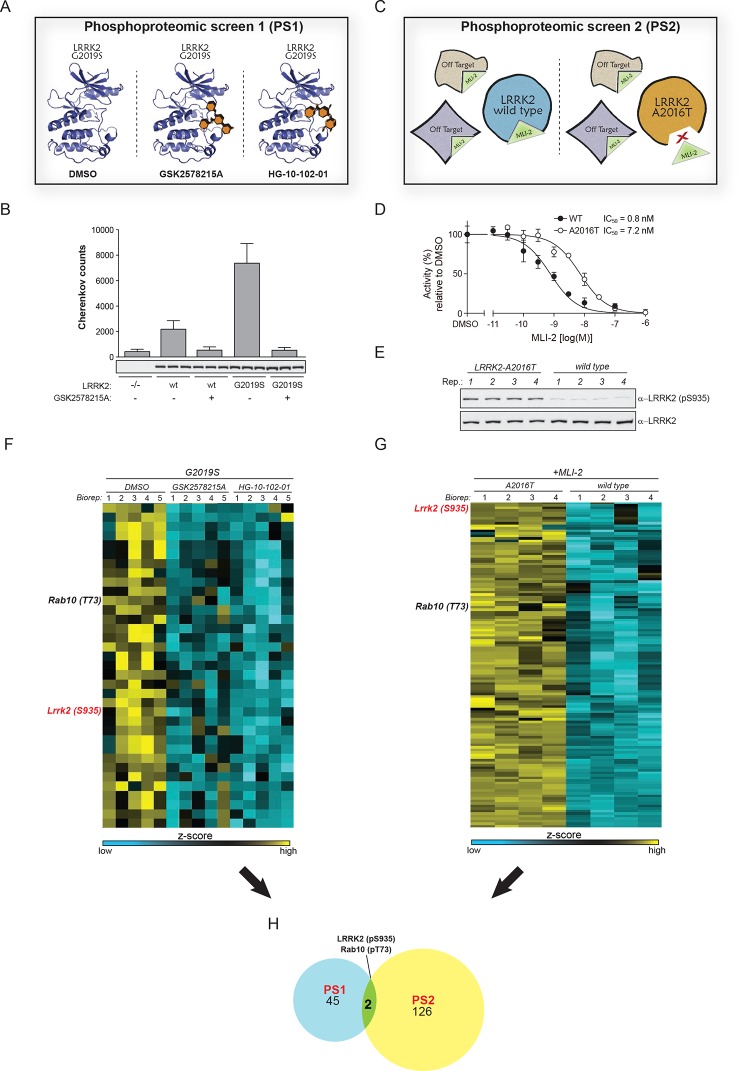
Figure 1. Two unbiased phosphoproteomic screens identify physiological LRRK2 targets.
(A) Experimental setup of PS1. LRRK2-G2019SGSK mouse embryonic fibroblasts (MEFs, n=5) were treated with DMSO or each of two structurally distinct LRRK2 inhibitors GSK2578215A or HG-10-102-01 (1 µM for 90 min). (B) LRRK2 immunoprecipitated from either knockout (-/-), wild-type (wt) or LRRK2-G2019SGSK (G2019S) knock-in MEFs was assessed for phosphorylation of Nictide (Nichols et al., 2009) peptide substrate in absence or presence of GSK2578215A (2 μM). Western blot below shows that similar levels of LRRK2 were immunoprecipitated. Error bars are mean ± SD (n=3). (C) Scheme of PS2. The higher affinity of MLI-2 toward wt-LRRK2 allows specific pinpointing of LRRK2 substrates when comparing the phosphoproteomes of wt and A2016T MEFs. (D) Kinase activities of wt (closed circles) and A2016T (open circles) GST-LRRK2 [1326-2527] purified from HEK293 cells were assayed in the presence of the indicated concentration of MLI-2 (n=3). (E) Decreased levels of pS935-LRRK2 in wt MEFs after treatment with 10 nM MLI-2. (F) Heat map cluster of phosphopeptides in PS1 (p<0.005) which are downregulated after treatment with both GSK2578215A and HG-10-102-01. (G) Heat map cluster of downregulated (FDR=0.01, S0=0.2) phosphopeptides in PS2. (H) Venn diagram of overlapping downregulated phosphosites in PS1 and PS2. (Biorep= biological replicate). SD, standard deviation.