Figure 4.
Scc3’s Highly Conserved Surface Is Essential for NScc1 Dissociation
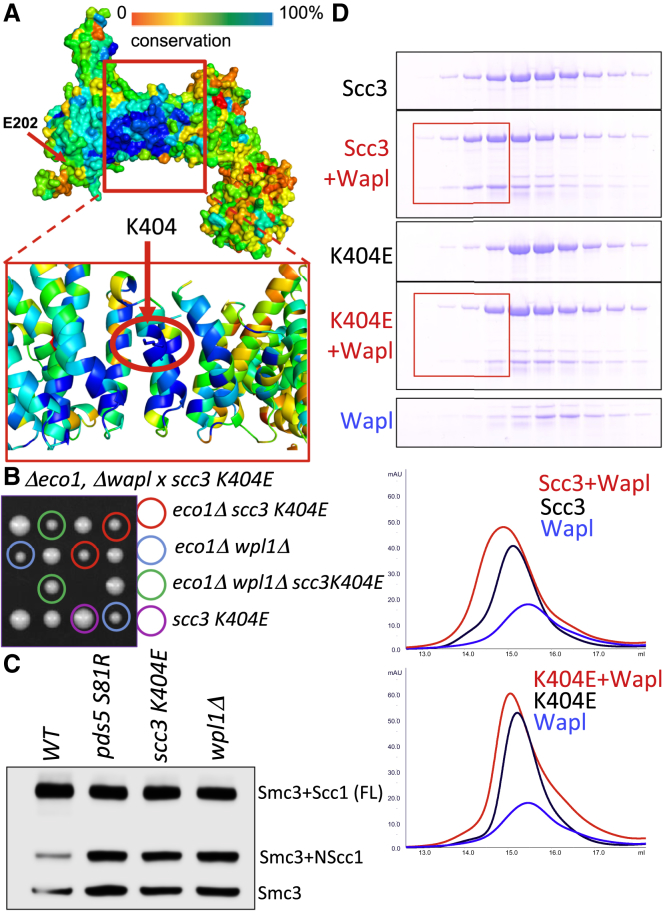
(A) Surface conservation of Scc3 orthologs projected on Z.r. Scc3 (blue, most conserved; red, least conserved) highlighting the conserved K404 (S. cerevisiae).
(B) Diploid strain (MATa/α wpl1Δ eco1Δ scc3K404E) was sporulated, tetrads dissected, and selected haploid segregants with their genotypes shown.
(C) Exponentially growing cells from wild-type (K22156), wpl1Δ (K22155), pds5-S81R (K20521), and scc3-E404K (K24349), all MATα SMC3(S1043C)-HA6 MYC3-SCC1(TEV268) were grown in YPD medium at 25°C and treated with 5 mM BMOE to crosslink Smc3 S1043C and Scc1 C56. Crosslinking was analyzed by western blotting using anti HA antibody.
(D) HIS-tagged wild-type Scc3, Scc3K404E, and Wapl proteins were purified from E. coli using TALON resin followed by Size exclusion using Superdex 200 16/60 column. Wild-type Scc3 or the K404E mutant protein was incubated either alone or with Wapl. After separation of the proteins by gel filtration using a Superose 6 column, the peak fractions were analyzed by SDS-PAGE and Coomassie staining, the fractions containing the Scc3/Wapl complexes are highlighted with a red box. The peak profiles of Scc3, Wapl, and the Scc3 Wapl complex are shown in the right for the wild-type and the E404K mutant proteins.