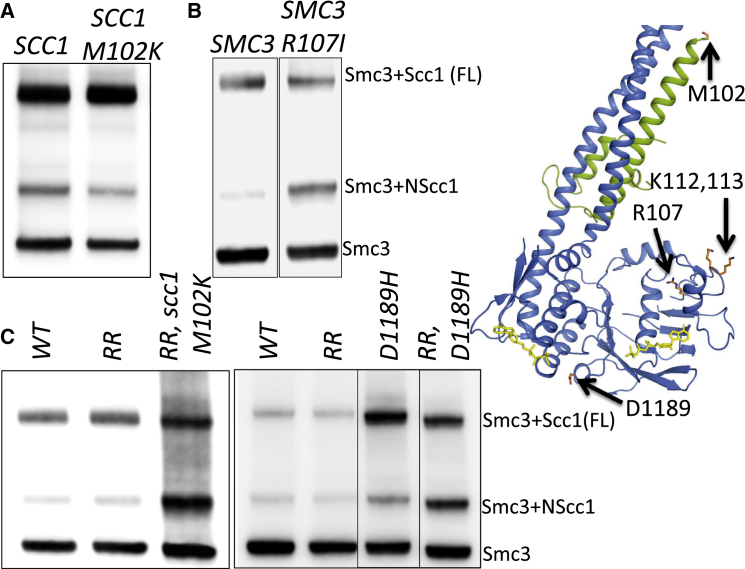
Figure 5.
Effect of smc3 and scc1 Mutations on NScc1 Release
(A) Strains K24297 (MATa SMC3(S1043C)-HA6 MYC3-SCC1) and K24343 (MATa SMC3(S1043C)-HA6 MYC3-scc1M102K) growing exponentially in YPD medium at 25°C were treated with 5 mM BMOE to crosslink Smc3 S1043C with either wild-type Scc1 or Scc1 M102K. The crosslinking was analyzed by western blotting using an HA epitope-specific antibody. The structure of Smc3-Scc1NTD complex (PDB: 4UX3) is shown on the right with Scc1 M102, Smc3 R107, K112, K113, and D1189 residues marked.
(B) Strains K24217 (MATa SMC3 URA3::SMC3 S1043C-PK6 MYC3-SCC1) and K24485 (MATa SMC3 URA3::SMC3 R107I S1043C-PK6, MYC3-SCC1) were treated as in (A) and crosslinking analyzed by western blotting using anti PK antibody. The data are from the same western blot, with irrelevant lanes removed.
(C) Strains K24217 (MATa SMC3 URA3::SMC3 S1043C-PK6 MYC3-SCC1), K24493 (MATa SMC3 URA3::smc3 K112 K113R S1043C-PK6 MYC3-SCC1), K24495 (MATa, SMC3, URA3::smc3 K112 K113R S1043C-PK6 MYC3-SCC1 M102K), K24497 (MATa SMC3 URA3::SMC3 S1043C D1189H-PK6 MYC3-SCC1), and K24499 (MATa SMC3 URA3::smc3 K112 K113R S1043C D1189H-PK6 MYC3-SCC1) were analyzed as in (B). The data shown in the right panel are from the same blot, with irrelevant lanes removed.