Fig. 1.
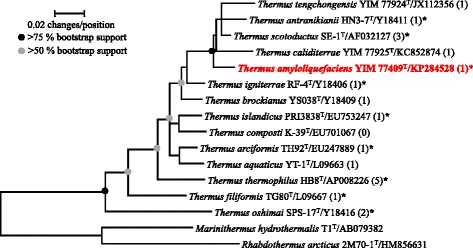
Maximum-likelihood phylogenetic tree of the genus Thermus to highlight the position of Thermus amyloliquefaciens strain YIM 77409T. The tree was reconstructed based on 1374 aligned positions that remained after the application of the Lane mask to the 16S rRNA gene sequences using MEGA 5.0 [54]. Complete deletion of gaps and missing data and Kimura’s two-parameter model was applied. Bootstrap analysis was based on 1000 resamplings. Nodes supported in >75 % (black circles) or >50 % (grey circles) of bootstrap pseudoreplicates (1000 resamplings) for both maximum-likelihood and neighbor-joining methods are indicated. Bar, 0.02 changes per nucleotide. The number of genomes available for each species is included in parentheses (see Table 5) and the asterisk indicates that the genome of the type strain is available. The 16S rRNA gene sequences from Marinithermus hydrothermalis T1T/AB079382 and Rhabdothermus arcticus 2 M70-1T/HM856631 were used as outgroups
