Figure 1.
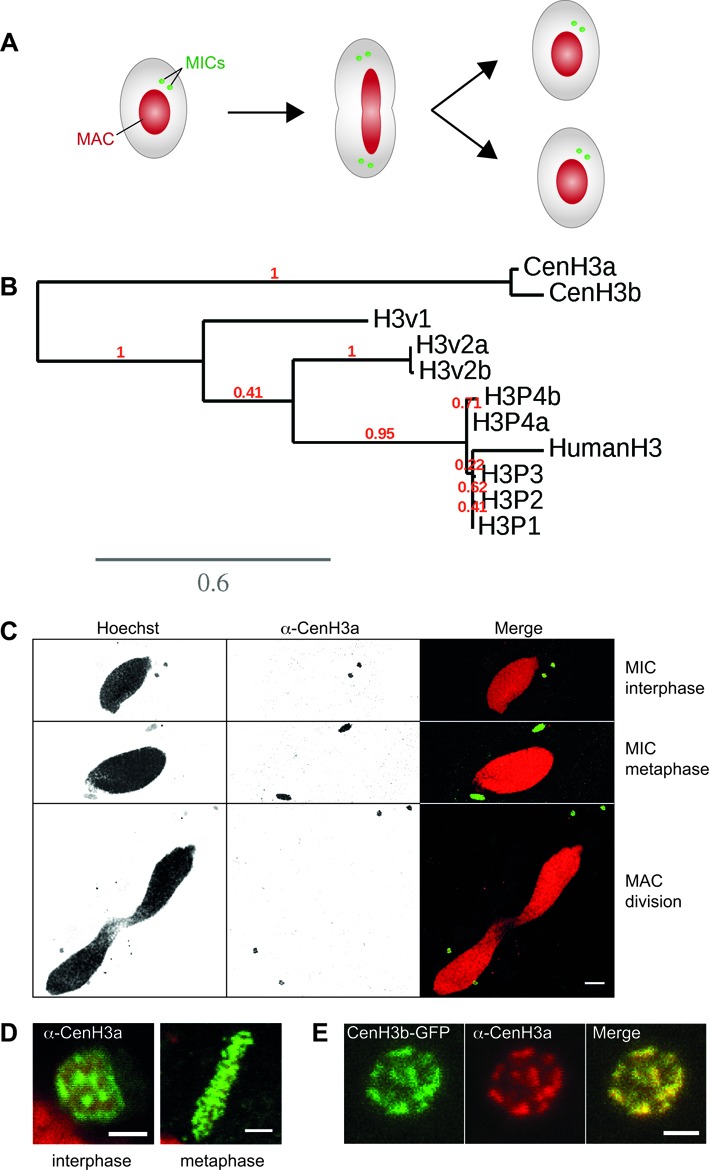
The Paramecium tetraurelia centromeric histone H3 variant. (A) Schematic representation of key nuclear events during Paramecium cell division. MAC: macronucleus; MICs: micronuclei. Note that the MICs divide before the MAC. (B) Phylogenetic analysis of P. tetraurelia H3 and H3 variants proteins. H3 proteins were retrieved using BLAST (55). Duplicates from the last whole genome duplication are named a and b. Multiple alignments were performed with the MUSCLE software (56). Phylogenetic analysis was carried out using PhyML 3.0 (bootstrapping procedure, 100 bootstraps) with default parameters and trees were visualized using TreeDyn (57). A scale bar in expected substitutions per site is provided for branch length. See also Supplementary Figures S1 and S2. (C) Immunostaining with CenH3a antibody at different stages of the cell cycle. Scale bar is 10 μm. (D) Magnified views of the MICs during interphase and metaphase. Scale bar is 2 μm. See also Supplementary Figure S3. (E) Colocalization of CenH3a and CenH3b proteins in the MICs during interphase. Immunostaining with CenH3a antibody of CENH3b-GFP transformed cells during vegetative growth. Scale bar is 2 μm.
