Figure 5.
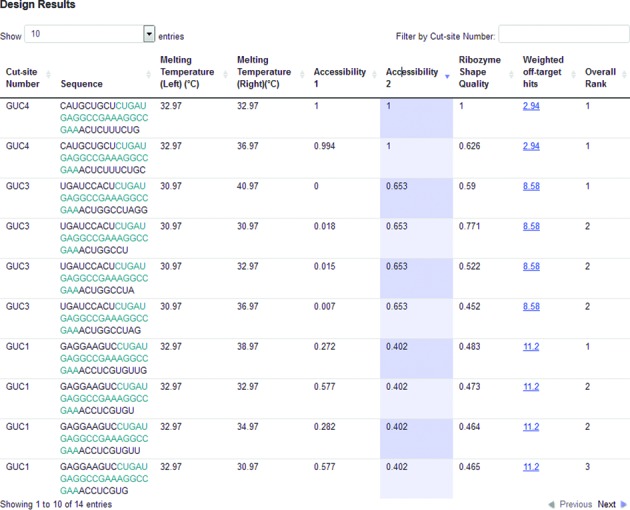
Sample output from RiboSoft. The cut-site number (starting at 0 for the first cut-site) and corresponding ribozyme sequence is shown for each candidate. All the parameters corresponding to this candidate are shown on the same row, Tm of both arms, accessibility (from 0, worst, to 1, the best), ribozyme shape (also from 0 to 1) and the overall rank. Off-target hits are shown only when the ribozyme was selected for in vivo use, 1 normally means that only the targeted gene is hit, higher numbers imply potential non-specific activity.
