Figure 8.
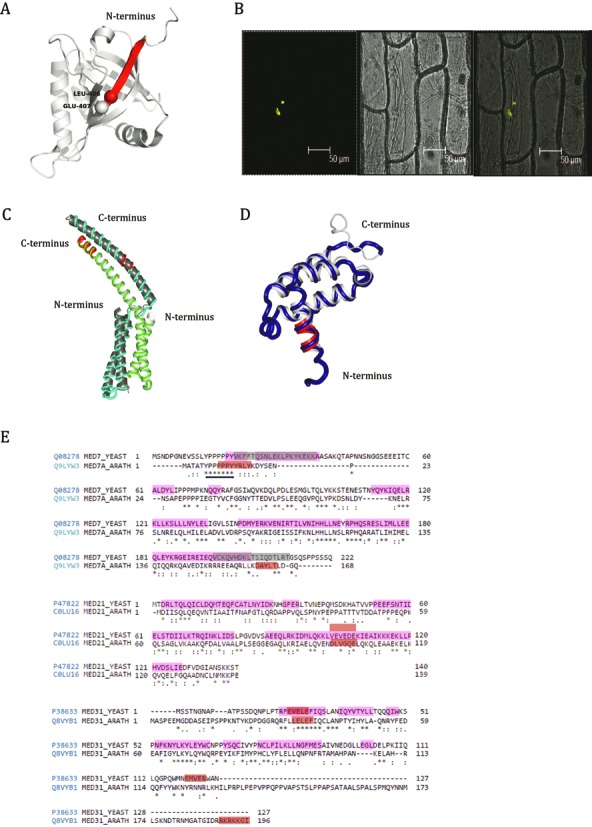
MoRFs in Mediator complex subunits of Arabidopsis, yeast and human. (A) Junction-MoRF in ACID domain of human Med25 is shown in red. Two Amino acids (L406 and Q407) involved in interaction with VP16 TADs are highlighted as spheres. Structure of ACID domain was generated using PDB id 2XNF. (B) Interaction between AtMed7 and AtMed21 was confirmed by BiFC assay. (C) Homology models of AtMed7 (green), AtMed21 (cyan) and (D) AtMed31 (blue) are aligned on known yeast counterparts (PDB id 1YKH and PDB id 3FBI in gray). MoRFs in all the structures are shown in red color. (E) Conserved MoRFs in Med7, Med21 and Med31 of yeast and Arabidopsis are shown as red and gray boxes in sequence alignment. Gray boxes represent MoRFs published earlier whereas red boxes represent MoRFs predicted in this study. Double headed arrow represents the highly conserved poly proline stretch flanking MoRF in Med7. Residues with helical propensity are highlighted in pink. Interestingly, some MoRFs are in these highlighted regions.
