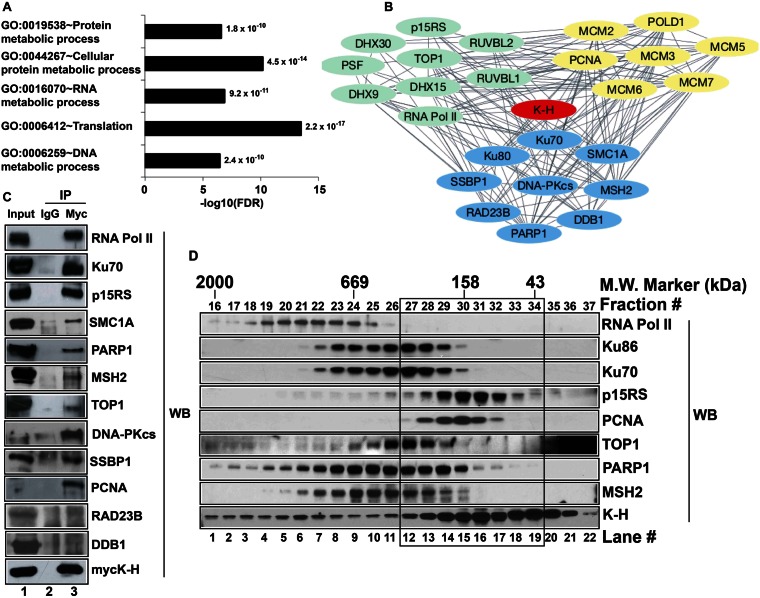
Figure 1.
Screening and validation of novel binding partners of K-H. (A) Identification of potential K-H binding partners was performed using TAP-MS screening. Functional annotation of identified proteins was determined by gene ontology (GO) analyses using DAVID v6.7. Bar graph shows the GO terms plotted against negative logarithms to the base 10 of FDR. Functional categories with –log10(FDR) cutoff value of >5 and indicated p-values are shown. (B) Select proteins were used to build a complex network of proteins associating with K-H using STRING 9.1 database and Cytoscape 3.1.0 software as described in ‘Materials and Methods’. Each single line connecting individual proteins represents a connection between given proteins. Proteins represented by green, blue and yellow colors are involved in RNA metabolism, DNA repair and replication, respectively. (C) Whole cell lysates prepared from 293T cells transiently expressing mycK-H were used for co-IP analyses. These cells were simultaneously co-transfected with unique siRNA-k-h oligomers specific for the 3′-UTR sequence of k-h mRNA to knockdown endogenous K-H expression levels. Antibodies to mycK-H or normal mouse IgG were then used to pull-down proteins and then Western blot analyses were performed to detect specific proteins using respective antibodies. Lane 1 represents input for IP and lanes 2 and 3 represent IP by IgG and anti-myc antibodies, respectively. Note that co-IP data validates 10/12 tested interactions for proteins of most interest. (D) Nuclease-treated whole cell lysates were used to fractionate native protein complexes containing K-H using gel-filtration chromatography as described in ‘Materials and Methods’. Indicated proteins were detected by Western blot analyses. Note protein complexes containing K-H in the black box that are separate from RNA Pol II-containing K-H protein complexes.