Abstract
Kissing-loop annealing of nucleic acids occurs in nature in several viruses and in prokaryotic replication, among other circumstances. Nucleobases of two nucleic acid strands (loops) interact with each other, although the two strands cannot wrap around each other completely because of the adjacent double-stranded regions (stems). In this study, we exploited DNA kissing-loop interaction for nanotechnological application. We functionalized the vertices of DNA tetrahedrons with DNA stem-loop sequences. The complementary loop sequence design allowed the hybridization of different tetrahedrons via kissing-loop interaction, which might be further exploited for nanotechnology applications like cargo transport and logical elements. Importantly, we were able to manipulate the stability of those kissing-loop complexes based on the choice and concentration of cations, the temperature and the number of complementary loops per tetrahedron either at the same or at different vertices. Moreover, variations in loop sequences allowed the characterization of necessary sequences within the loop as well as additional stability control of the kissing complexes. Therefore, the properties of the presented nanostructures make them an important tool for DNA nanotechnology.
INTRODUCTION
Two single-stranded nucleic acid strands (either RNA or DNA) can anneal to each other via Watson–Crick base pairing if the two strands have a region of complementary bases. In canonical annealing, at least one strand has a free end and can therefore wrap around the other strand, generating a helical structure. An alternative form of hybridization is when two closed stem-loop nucleic acid sequences without a free 3′ and 5′ end hybridize via their single-stranded loop region, which is called kissing-loop annealing. This phenomenon was first described for RNA–RNA loops and was later extended to DNA–DNA loops (1,2). RNA–RNA kissing interaction is especially of great biological importance, e.g. as an intermediate step in the dimerization of the RNA genomes of the human immunodeficiency virus (HIV) (3) and the hepatitis C virus (4). Due to the medical importance of this RNA–RNA interaction, a series of investigations of the structure and properties of these kissing complexes have been performed (5–7). In contrast, knowledge about DNA–DNA kissing complexes is still rudimentary. DNA–DNA kissing complexes have been proposed as fuel for DNA devices (e.g., DNA nanobots) because if the sequences of the stem-loop structures are adequately designed, the DNA–DNA kissing complex is metastable and can be converted into a regular, more stable DNA duplex, which has much lower free energy. Thus, both in nature and in nanotechnology so far, the kissing-loop is only an intermediate on the path to a classical nucleic acid-nucleic acid duplex. In contrast, we chose an approach in which the DNA–DNA kissing complex cannot further hybridize into a longer duplex. This was achieved by adding stem-loop sequences to the vertices of DNA tetrahedrons and will preserve the reversibility of the kissing-loop complex because only the short stretch of hybridized nucleotides in the kissing complex has to be melted into two single loops.
The DNA tetrahedrons that were first designed by Goodman et al. (8) in 2004 were very stable against environmental influences and physiological conditions, as has been demonstrated in living mammalian cells and in blood (9,10). These tetrahedrons therefore represent promising structures for nanotechnological applications, such as use as a shuttle for drugs (11,12) or as an imaging material for tissue analysis (13). Furthermore, the assembly of tetrahedrons is relatively simple (14–17). Each tetrahedron can be modified in terms of size and the relative lengths of the edges (14). Further modifications include covalent linkage or reversible binding of molecules such as nanogold (15) or proteins to the tetrahedron, without interfering with its assembly (16,17). However, the application of DNA tetrahedrons might be increased if it was possible to build a complex of several tetrahedrons. This concept has only been realized by unspecific accumulation (18–20) and linear complex formation (21). In this study, we functionalized the tetrahedrons with specific stem-loop sequences located at the vertices of the tetrahedron to allow specific complex formation enabled by kissing-loop interaction of the single-stranded loops. We also characterized the influence of different factors, like the favored hybridizing sequences within the loop, the ionic concentration and the temperature of the surrounding milieu, on the yield and stability of the resulting complexes.
MATERIALS AND METHODS
Construction of functionalized DNA tetrahedrons
DNA oligonucleotides were synthesized; 5′-phosphorylated, if required (for the construction of covalently closed tetrahedrons); and purified by polyacrylamide gel electrophoresis (PAGE) by biomers.net GmbH, followed by dissolution in deionized water to attain a concentration of 100 μM. The sequences of the oligonucleotides used in this study are listed in Supplementary Table S1. They combine sequences leading to the stem-loop parts of the tetrahedrons (22–24) with sequences leading to a rigid DNA tetrahedron scaffold (14), linked by two adenine nucleotides that do not hybridize in the final structure. These newly combined sequences were checked for unwanted secondary structures using the Nupack web server (available at http://www.nupack.org/).
Preparation of functionalized DNA tetrahedrons
Tetrahedrons were built by mixing the four composing oligonucleotide strands to a final concentration of 2 μM each and by annealing via heating to 95°C for 2 min and fast cooling (within 1 min) to 4°C in TAE/Mg2+ buffer (40 mM Tris, 19 mM acetic acid, 1 mM EDTA and 12.5 mM magnesium acetate, pH 8 (22)). If ligation was required, NEB Quick Ligation Reaction Buffer was added for the annealing and T4 DNA ligase (Thermo Scientific, 0.5 Weiss units/μl) afterward for the enzymatic reaction. After incubation for 1 h at 16°C, Proteinase K (Carl Roth, 2 μg/μl) was added for 30 min at 37°C.
Kissing-loop interaction between functionalized DNA tetrahedrons
If not otherwise specified, loop–loop hybridization reaction was performed on ice or at room temperature by mixing equimolar quantities of tetrahedrons to a final tetrahedron concentration of 2 μM in TAE/Mg2+. To determine the influence of different cations on kissing-loop stability, TA buffer (40 mM Tris and 19 mM acetic acid) was used with or without the addition of magnesium chloride (Mg2+), calcium chloride (Ca2+) or zinc chloride (Zn2+) instead of TAE/Mg2+ buffer for tetrahedron assembly and the kissing reaction. Ligated tetrahedrons were mixed to a final concentration of 1 μM in TAE/Mg2+.
Native gel electrophoresis
Kissing complexes and monomeric tetrahedrons (as controls) were analyzed by native PAGE using a gel-running buffer identical to the respective hybridization buffer. The sample buffers (3×) for native gel electrophoresis contained 25% glycerol, 0.2% bromophenol blue, 0.2% xylene cyanol FF, 50 mM Tris–HCl (pH 8), and respective bivalent ions at the final indicated concentration. Gels were stained by silver staining or with GelStar® Nucleic Acid Gel Stain (Lonza). For the latter fluorescence staining, the GelStar® was diluted 10 000× in deionized water. The silver-staining protocol was modified according to Sanguinetti et al. (25) (Supplementary Table S2). To characterize the thermal stability of different kissing complexes, gels were run at different temperatures, as adjusted by circulating water through the gel chamber (Whatman Multigel-Long, Biometra) and a thermostat. For statistical evaluation, the staining intensity was quantified by using ImageJ image analysis software (available at http://imagej.nih.gov/ij) and the AIDA Image Analyzer (Raytest).
RESULTS
A DNA tetrahedron can be mono-functionalized at a vertex by stem-loop sequences
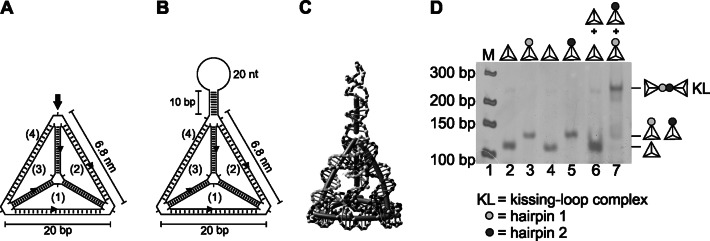
The properties of DNA allow the construction of various structures, including 3-dimensional (3D) ones, in addition to the well-known DNA helix. A principle underlying the building of such non-canonical structures is that base pairing is not continuous; rather, strands cross each other, or the DNA is bent to allow complementary base pairing in a different spatial orientation, e.g. with a different strand. This is the case not only for naturally occurring Holliday junctions (26) or artificial DNA origamis (27–29) but also for DNA tetrahedrons. Whereas the edges are formed by a DNA double helix, three short single-stranded regions are present at the vertices. In the case of a previously successfully built DNA tetrahedron (14), those three single-stranded regions were composed of only a single adenosine (Figure 1A). In the present study, we wanted to exploit the possibility of functionalizing the vertices of such tetrahedrons with stem-loop sequences. Stem-loops are very common in living systems and are found in single-stranded DNA or, more often, in RNA molecules. The so-called kissing-loop base pairing between two adequately designed loops should control the interaction of DNA tetrahedrons; for this reason, the stem region was chosen to be long enough (10 bp) to prevent denaturation of the tetrahedron caused by mechanical strain based on hybridization, as shown for RNA kissing of hairpins (30). To start simple, we asked whether it is possible to modify one strand to yield a DNA tetrahedron with one attached stem-loop at one vertex (Figure 1B and C). This structure was indeed built by annealing of four oligonucleotides. The functionalized DNA tetrahedron migrated as a single band in a native gel, with slower migration than that of the unfunctionalized tetrahedron (Figure 1D, lane 3 versus lane 2). By using equimolar amounts of all four strands, the structure assigned as a tetrahedron was formed, with high yield. If this formed structure was covalently closed by ligation using 5′-phosphorylated oligonucleotides and T4 DNA ligase, it was resistant to digestion with exonuclease III (ExoIII) (the structures shown in Figure 1D were treated in this way), demonstrating that the tetrahedron was correctly assembled. It was also possible to form the mono-functionalized tetrahedron with a different sequence in the loop region (Figure 1D, lane 5).
Figure 1.
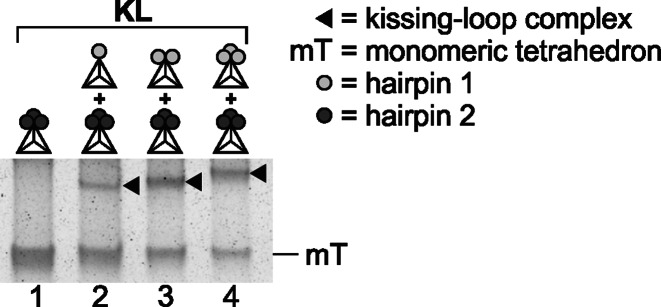
Tetrahedrons and mono-functionalized tetrahedrons. (A) Schematic drawing of an unfunctionalized DNA tetrahedron composed of strands (1–4), highlighting the unpaired nucleotides present in all strands at the vertices. (B) Schematic drawing of a mono-functionalized DNA tetrahedron with a stem-loop element (of 10 bp and 20 nt) integrated into strand (4), with two flanking unpaired nucleotides (adenines). (C) 3D simulation of the mono-functionalized tetrahedron shown in (B), performed with the open-source 3D modeling and simulation program NanoEngineer-1 (Nanorex, Inc.). (D) Native 6% PAGE (silver stain) analysis of the unmodified and two different mono-functionalized tetrahedrons (after ligation and ExoIII treatment). Whereas the stem sequence is the same for all stem-loop elements, two complementary loop sequences (1 and 2) exist, as represented by differently shaded circles. The kissing-loop (KL) complex formed by tetrahedrons with complementary loops in the stem-loop elements in the presence of 11.5 mM Mg2+ is schematically shown on the right.
Two mono-functionalized DNA tetrahedrons interact as a kissing-loop complex
The stem-loop DNA sequences of the two different DNA tetrahedrons described above were designed in such a way that both tetrahedrons could assemble by kissing-loop interaction. The two designed loop sequences were selected to be 20 nt long and completely complementary for improved kissing interaction, and the stem-forming sequence was the same for all stem-loop sequences (hairpin 1 and hairpin 2). This is schematically indicated by different shading in the figures (dark and light gray), and a detailed drawing is shown in the Supplementary Material (Supplementary Figure S1). Whereas the incubation of unfunctionalized tetrahedrons did not lead to dimeric or multimeric complex formation, as indicated by native gel electrophoresis, (Figure 1D, lane 6), we could indeed demonstrate effective kissing interaction after mixing the two different mono-functionalized tetrahedrons, as indicated by a shift in electrophoretic mobility (Figure 1D, lane 7).
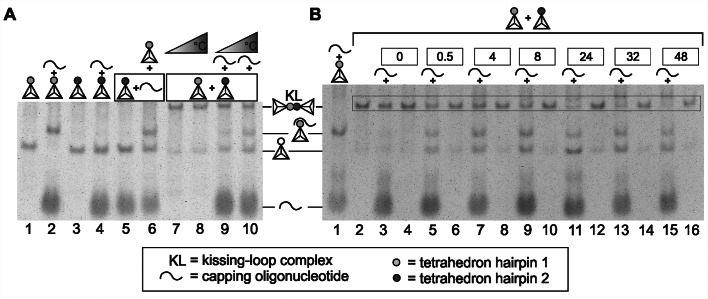
Disassembly of kissing complexes by competition
For DNA hairpins, it has been proposed that the distribution of the four different nucleobases influences the structure of a formed kissing complex (23,24). Here, we used a loop with separated AT- and GC-rich regions, which should allow lateral kissing interaction (24). Irrespective of the structure of the kissing complex, complete hybridization of the loop sequence was predicted not to be possible, as several nucleotides would remain unengaged in base pairing. Therefore, we wanted to analyze the influence of the presence of free single-stranded DNA that was fully complementary to one of the two DNA loop sequences. In particular, we added a capping oligonucleotide composed of a tandem repeat of the sequence complementary to the 20-nt loop region of hairpin 1, separated by 40 thymidine nucleotides. As expected, the addition of this capping oligonucleotide to the tetrahedron with hairpin sequence 1 led to the appearance of a species with a higher molecular weight, whereas the free tetrahedron disappeared (Figure 2A, lanes 1 and 2). Addition of the capping oligonucleotide to the tetrahedron with hairpin sequence 2 did not alter the electrophoretic mobility of this tetrahedron, indicating the absence of an interaction of this capping oligonucleotide with loop sequence 2, as expected (Figure 2A, lanes 3 and 4). When the capping oligonucleotide was added during the hybridization reaction, performed by mixing the first tetrahedrons with hairpin 2 with the capping oligonucleotide and then adding this mixture to the tetrahedrons with hairpin 1, kissing-loop complex formation between the two tetrahedrons was impaired (Figure 2A, lane 6). Therefore, we next asked whether the addition of the capping oligonucleotide would interfere with a pre-built kissing-loop complex as well. We tested the disintegration of the tetrahedron kissing complex via the addition of the capping oligonucleotide on ice and at 25°C. Whereas the kissing complex was stable on ice, we could measure a 21% decrease in the amount of the kissing complex after a temperature increase for 25 min (Figure 2A, lanes 7–10). In a further experiment we analyzed the slow disintegration at 25°C in detail (Figure 2B). For this we extended the incubation time of capping oligonucleotide and kissing complex up to 48 h. Interestingly the main fraction of kissing complex was not disintegrated before 24 h incubation time. To investigate if the kissing complex and the tetrahedron-capping oligonucleotide are in an equilibrium, we incubated the complex of capping strand hybridized to tetrahedron hairpin 1 with tetrahedron hairpin 2 over 48 h, but the complex remained stable and no kissing complex was detected (Supplementary Figure S2). This might be an interesting effect in view of the possible application of kissing DNA tetrahedrons as a shuttle; the slow degradation of a kissing complex by the invasive strand might enable the slow release of potentially encapsulated molecules, as discussed later.
Figure 2.
The targeted disassembly of a kissing complex is triggered by the invasion of a capping oligonucleotide complementary to the loop sequence. (A) The kissing complex and the constituting tetrahedrons were incubated for 25 min with a 2-fold excess of capping oligonucleotide with complementarity to loop sequence hairpin 1. In the case of pre-incubation, the respective molecules are indicated by a box. Schematic drawings of the structures visible after native gel electrophoresis on 4% polyacrylamide gels (with 11.5 mM Mg2+) are indicated on the right. The experiment was performed at two different temperatures: on ice (lanes 7 and 9) or at 25°C (lanes 8 and 10). Tetrahedrons with empty circles indicate the position of monomeric tetrahedrons, irrespective of the nature of the loop sequence (1 or 2). (B) In an extended experiment at 25°C, it was possible to show the disintegration of the kissing complex (lane 2) as a slow one-way reaction over 48 h (0–48, lanes 3–16) by formation of a complex of tetrahedron hairpin 1 and capping strand (lane 1). The reaction starts at time point 0 h by addition of the capping oligonucleotide whereas a control sample without capping oligonucleotide was analyzed for each time point, too. Due to the slow kinetics the major fraction of kissing complex was only disassembled after more than 24 h.
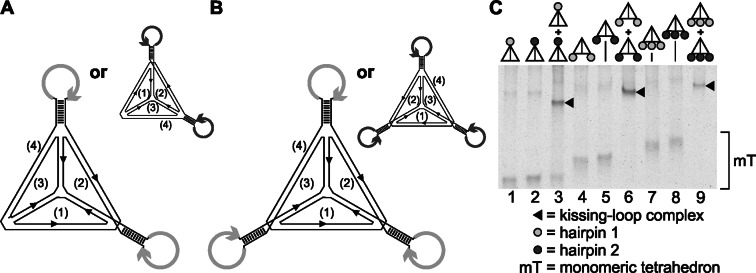
A DNA tetrahedron can be modified at multiple vertices by stem-loop sequences, and such molecules form kissing complexes
The design of the tetrahedrons might also allow the functionalization of multiple vertices of one tetrahedron with stem-loop sequences. We decided to design tetrahedrons with one, two or three functionalized vertices and integrated the stem-loop sequences into different tetrahedron-composing strands, so that the resulting tetrahedrons would look as schematically shown in Figures 1B and 3A and B. As described above, two types of stem-loop-forming sequences were used. Each tetrahedron was designed to contain either only loop sequence 1 or only loop sequence 2 (light and gray circles).
Figure 3.
Tetrahedrons with two or three stem-loops at their vertices can be assembled to form kissing complexes. (A and B) Schematic drawings of the tetrahedrons. Whereas the stem sequence is the same for all stem-loop elements, two complementary loop sequences (1 and 2) exist, as represented by differently colored circles (dark and light arrows) (Supplementary Figure S1). (C) Different DNA tetrahedrons, as schematically shown at the top of the lanes, were hybridized in the presence of 11.5 mM Mg2+ (representative result of at least three independent repetitions). Dimeric tetrahedron complexes are indicated by black triangles. Separation of monomeric tetrahedrons and kissing complexes was performed with a 4% polyacrylamide gel and was visualized by fluorescence staining.
We were indeed able to assemble tetrahedrons with one, two or three functionalized vertices. Those structures migrated as a single band in a native gel, with decreasing electrophoretic mobility as the number of functionalized vertices increased (Figure 3C, lanes 1 and 2, 4 and 5, and 7 and 8). After ligase treatment, the structures were stable against exonuclease digest, demonstrating that all strands were ligated, which should only be possible if the tetrahedral structure is formed (Supplementary Figure S3). Next, we asked whether an interaction between the multi-functionalized tetrahedrons with loop sequences 1 and 2 is possible. As shown by native gel electrophoresis, the tetrahedrons functionalized at multiple vertices interacted (Figure 3C, lanes 3, 6 and 9). We were also able to establish tetrahedrons with further loop sequences (loop sequence hairpin 3 and the complementary loop sequence hairpin 4) that could hybridize with an identical kissing efficiency (Supplementary Figure S4A). Furthermore we could demonstrate kissing interaction of multifunctionalized tetrahedrons that exhibit a combination of different loop sequences. For this purpose we designed two tetrahedrons with two loops each, one functionalized with both hairpin 1 and hairpin 3 loop sequences and the other one with loop sequences hairpin 2 and 4 (Supplementary Figure S4B).
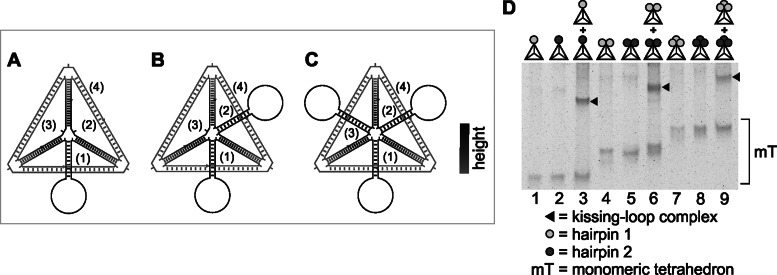
A DNA tetrahedron can be multi-functionalized at one vertex with up to three stem-loop sequences
To further evaluate the potential of our approach, we designed tetrahedrons with two or three stem-loops located at the same vertex (Figure 4). As before, different tetrahedrons with complementary loop sequences were designed. Again, all stem-loop sequences were part of separate DNA strands with sequence directionality to allow multiple kissing events. The results indicate efficient formation of a distinct kissing complex for each tested tetrahedron incubated with its partner (Figure 4D). After having demonstrated the feasibility of our approach, we continued with an in-depth analysis.
Figure 4.
Tetrahedrons with two or three stem loops at the same vertex can be assembled to form kissing complexes. (A–C) Schematic drawings of the tetrahedrons with one stem-loop (A), two stem-loops (B) or three stem-loops (C) added to the same vertex, as observed from the top. (D) The use of two different loop sequences (represented by differently shaded circles) led to two complementary types of tetrahedrons (representative result of at least three independent repetitions). Interaction was analyzed in the presence of 11.5 mM Mg2+ by native 4% PAGE and fluorescence staining. Dimeric tetrahedron complexes are indicated by black triangles.
The loop number and position influence kissing interaction and complex stability
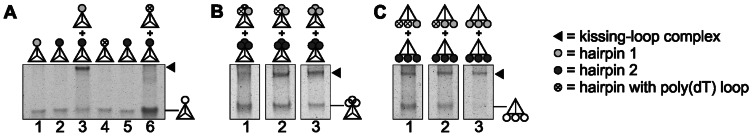
We next asked about the factors influencing the stability of kissing complexes. We first focused on the impact of the total number of loops per tetrahedron involved in kissing interaction. We started with loops positioned at one vertex of the tetrahedron and analyzed kissing-loop interaction between a tetrahedron with three loops and tetrahedrons with the complementary loop sequence and one, two or three stem-loops (Figure 5). Although we already showed (Figure 4) that the three-looped tetrahedrons with complementary loop sequences hybridized, a priori prediction of whether an increased number of loops at one vertex would increase the complex stability or weakens the interaction was difficult. The presence of the three loops could lead to steric hindrance due to crowding at the vertex or to an enhancement of kissing interaction due to a higher concentration of hybridization partners in the same area. Interestingly, kissing complexes formed between the three-loop tetrahedron and the one-, two or three-looped tetrahedron with the complementary loop sequence. However, an improved conversion from monomer to kissing complex was observed when complementary loops were present in similar numbers, as in the two complementary three-loop tetrahedrons. We wanted to explore this effect in detail and thus continued working with tetrahedrons with three loops at the same vertex. However, we took advantage of a loop sequence consisting of 20 thymines ((dT)20). Tetrahedrons with such functionalization did not form a kissing complex with tetrahedrons with loop sequence 2 (Figure 6A). We substituted loop sequence 1 with poly(dT) loops, which resulted in varying numbers of complementary loops, whereas the total number of loops at the tetrahedron remained constant, and we tested kissing interaction with the same tetrahedron as before (Figure 6B). Interestingly, kissing complexes with the same electrophoretic mobility could be observed in all different combinations. Based on the migration distance in comparison with that of DNA markers and based on the fact that the kissing product, which involves only one pair of complementary loops, has to be a dimer, all kissing products in this experiment were also dimers. This is in line with the relatively small changes in electrophoretic mobility for different kissing-loop products originating from tetrahedrons with different numbers of loops, as observed before, demonstrating that all kissing products in this study were dimers of tetrahedrons. We next wanted to compare this hybridization involving tetrahedrons tri-functionalized at one vertex (Figure 6B) with kissing-loop interaction between tetrahedrons with three loops, each at a different vertex (Figure 6C). In both cases, the fraction of tetrahedrons involved in detectable kissing-loop interaction increased with the number of complementary loops and fewer substitutions with poly(dT) sequences. However, there was a difference in hybridization efficiency between tetrahedrons that had three stem-loops at the same vertex and tetrahedrons with loops located at different vertices. In comparison, in time-limited experiments, 53% of kissing product was measured for hybridization at one vertex and 17% was examined for hybridization of two three-loop tetrahedrons at different vertices. Such an effect of the position of the loops (at the same or different vertices) was also detectable with the tetrahedrons with poly(dT) loops. If the poly(dT) loop(s) was/were positioned at the same vertex as the complementary loop(s), less distinct product bands, accompanied by a detectable smear in the lanes, were observed, indicating a less stable interaction (Figure 6B versus C, lanes 1 and 2). Interestingly, tetrahedrons with no poly(dT) loops (Figure 6A, lane 3) showed a more distinct dimer band and a higher band intensity than the hybridization of tetrahedrons possessing two additional poly(dT) loops (Figure 6B,C, lane 1). Thus, the vicinity of stem-loops, as in the case of multi-functionalization at the same vertex, can improve the hybridization efficiency (faster hybridization and more stability during electrophoresis) if these loops can participate in kissing interaction. Otherwise, non-complementary loops lead to destabilization of kissing interaction, rendering this system tunable for applications.
Figure 5.
Kissing complexes arise from tetrahedrons with varying number of loops at the same vertex. Gel analysis of the formation of kissing complexes with respect to yield between one-, two- and three-loop tetrahedrons (light gray circles) and the complementary three-loop tetrahedron (dark gray circle) by native 8% PAGE in the presence of 11.5 mM Mg2+ (representative result of at least three independent repetitions). The dimeric tetrahedron complexes are indicated by black triangles.
Figure 6.
The influence of additional non-complementary poly(dT) stem-loops on the tetrahedrons on kissing-loop formation. (A) Analysis of the possibility of kissing-loop interaction (lane 6) between a mono-functionalized tetrahedron with loop sequence 2 (dark gray, lane 5) and a mono-functionalized tetrahedron with a stem-loop sequence containing a (dT)20 loop (crossed circle, lane 4). As a control, kissing-loop formation by mono-functionalized tetrahedrons with loop sequences 1 (light gray) and 2 (dark gray) is shown (lanes 1–3). (B) Analysis of kissing-loop formation and the stability of the respective kissing complexes formed between a tetrahedron tri-functionalized at one vertex with loop sequence 2 and tri-functionalized tetrahedrons with one, two or three loop sequences (loop sequence 1) and two, one or zero (dT)20 loop sequences, respectively. (C) Analysis of kissing-loop formation by a tetrahedron with loops located at three different vertices. Similar to (B), a tri-functionalized tetrahedron with loop sequence 2 was hybridized to three tetrahedrons with one, two or three loop sequences (loop sequence 1) and respective substitution of loop sequence 1 with one or two (dT)20 loop sequences. The migration of dimeric tetrahedron complexes is marked by black triangles. The tetrahedrons with empty circles on the right side of the images indicate the position of monomeric tetrahedrons, irrespective of the nature of the loop sequence (1, 2 or (dT)20). All samples were analyzed by native 8% PAGE in the presence of 11.5 mM Mg2+ (representative result of at least three independent repetitions).
Kissing complex stability at various temperatures
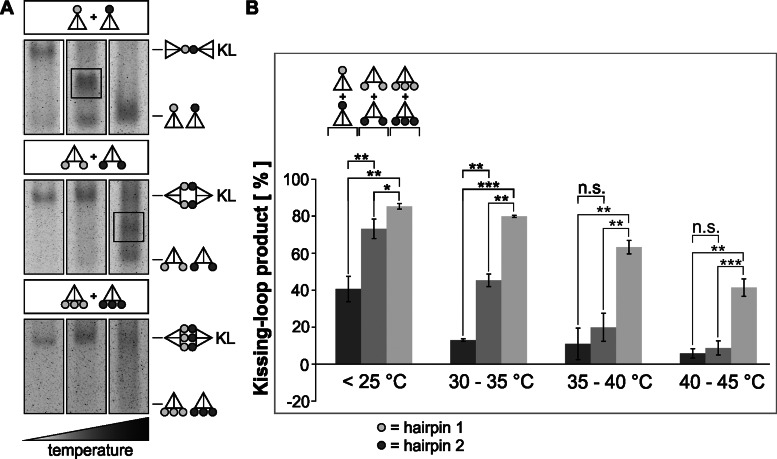
The interaction of two tetrahedrons via three loops at different vertices opens the possibility of using this complex of tetrahedrons as a biocompatible shuttle for pharmaceutical applications. In the following experiments, we wanted to analyze further factors that influence the stability of kissing complexes. Temperature changes can exhibit stabilizing or destabilizing effects on DNA double strands, influencing the coaxial stacking within the helix in a sequence-dependent manner (31). Similar effects are also observed for kissing complexes (32). At room temperature, the HIV-1 RNA kissing complex is formed in a thermodynamically metastable state, and coaxial stacking stabilizes the complex, which can only be disrupted by binding of the protein NCp7 or artificial induction by increasing the temperature (33). We could achieve the formation of a DNA kissing complex at 25°C. To analyze the effect of temperature on kissing tetrahedron dimers, temperature assays were conducted below 25°C, between 30°C and 35°C, and at physiologically relevant temperatures (from 35°C to 40°C and from 40°C to 45°C). The tested complexes were formed between two tetrahedrons with one loop each, two tetrahedrons with two loops and two tetrahedrons with three loops. Figure 7 demonstrates the effect of increasing the temperature on the disintegration of kissing complexes to form monomeric tetrahedrons, as indicated by a band shift from kissing complex to monomer. The monomeric band intensified at increased temperatures, whereas the kissing complex band faded. This effect was diminished for complexes with higher numbers of loops, and the kissing complex formed by two tetrahedrons with three loops each showed the highest persistence against a temperature increase. Thus, we could demonstrate a dependency between loop numbers, which heighten the probability of forming several kissing-loops, and the resulting temperature resistance. Interestingly, a temperature increase led to the formation of an intermediate before total disintegration at higher temperatures (Figure 7A, rectangle).
Figure 7.
The temperature stability of the DNA kissing complex depends on the number of stem-loops. Three different sets of KL complexes of tetrahedrons with varying numbers of loops positioned at different vertices were analyzed on native 4% polyacrylamide gels containing 11.5 mM Mg2+ at temperatures ranging from below 25°C to 45°C. (A) Analysis of the electrophoretic migration behavior of the mixture of structures present at below 25°C (left lane), at 30–35°C (middle lane) and at 35–40°C (right lane) for the given kissing dimers. Intermediate structures are boxed. (B) The percentage of stable kissing complexes was calculated by measuring the changes in the band intensity of the kissing complex and the monomer at rising temperatures. The mean values (bars), including the standard deviation (error bars) of at least three independent measurements, are depicted. The asterisks indicate the significance level: t-test, *P < 0.05, **P < 0.001 and ***P < 0.0001.
Kissing complex stability at various ionic concentrations
The omission of Mg2+ during gel electrophoresis of kissing complexes formed in the presence of Mg2+ led to disintegration of the kissing complexes. Monovalent K+ was not (at least at an equimolar concentration) an adequate substitute for the bivalent ion Mg2+ (Supplementary Figure S5). However, only Mg2+ was used in previous DNA–DNA kissing-loop studies (2,22,34). Therefore, we considered the effect of other bivalent cations, namely, Ca2+ and Zn2+. Again, we used complementary tetrahedrons with one, two or three loops at different vertices and used different concentrations of Mg2+, Ca2+ and Zn2+ during kissing-loop formation and gel electrophoresis (Figure 8). Interestingly, kissing complexes could be formed with Mg2+, Ca2+ and Zn2+. However, as was the case when analyzing the effect of a temperature increase (Figure 7), this analysis also showed fewer kissing-loop products when less loops were available for a possible interaction. This was observed with all three cations at all three tested concentrations (0.625, 1.25 and 11.5 mM) (Figure 8). Additionally, effects on kissing-loop stability provoked by the tested three ions (Mg2+, Ca2+ and Zn2+) were observable and were especially prominent under defined conditions. Whereas at high ionic concentrations, it could be concluded that the one-loop tetrahedron complexes are best stabilized by magnesium ions, rather than Ca2+ ions and Zn2+ ions, at low cation concentrations, a slightly more favorable effect of Mg2+ ions could be deduced for the di- and tri-functionalized tetrahedrons. A low bivalent cation concentration had an especially strong impact on kissing-loop stability in the case of Zn2+. No distinct kissing product band was observed for the one-loop tetrahedrons at 0.625 mM Zn2+, and only faint bands were visible on polyacrylamide gels for the two-loop tetrahedrons and the three-loop tetrahedrons at the same Zn2+ concentration. A kissing product band for the one-loop tetrahedron was first distinguishable at 1.25 mM Zn2+ (Supplementary Figure S6). In summary, we could demonstrate higher resistance against changes in the ionic concentration for complexes formed with tetrahedrons with increased numbers of loops that can be involved in kissing interaction.
Figure 8.
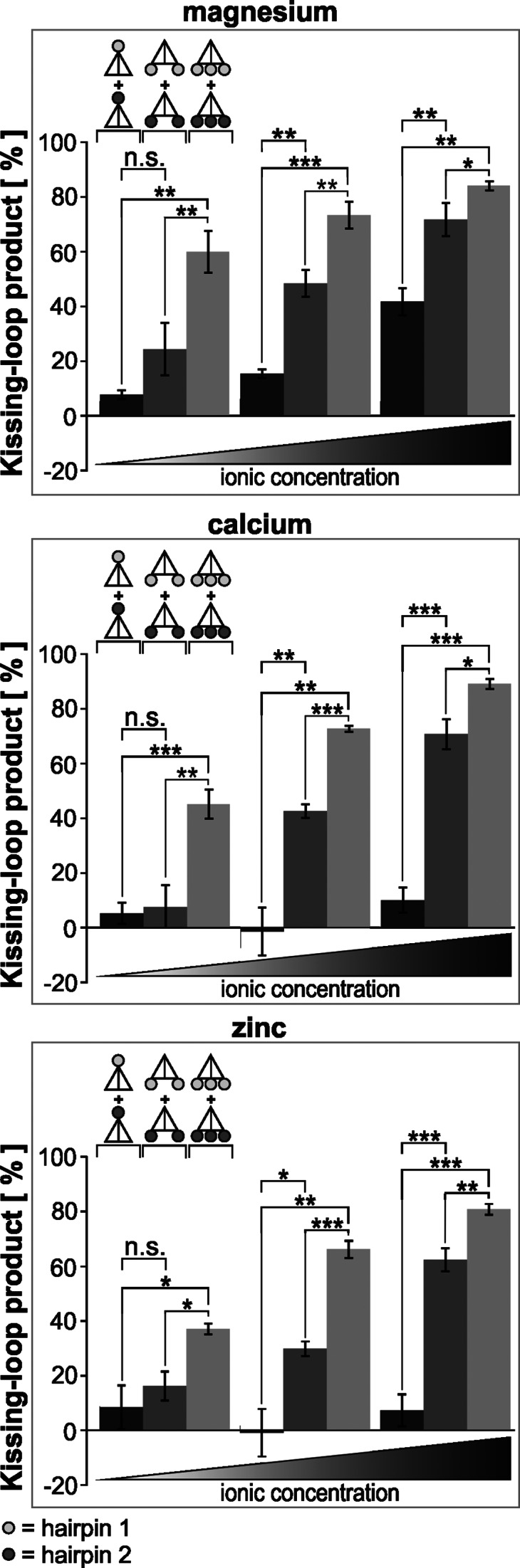
The kissing complex stability at various concentrations of bivalent cations depends on the number of possibly interacting loops. Kissing interaction was performed between two complementary types of tetrahedrons with one, two or three loops at different vertices in the presence of increasing concentrations of different ions. All gel electrophoretic analyses were performed below 25°C. The two types of complementary loops are represented by light gray or dark gray circles. The tested cation concentrations were 0.625, 0.125 and 11.5 mM (from left to right). The yield of kissing product as a percentage was determined by measuring the band intensity of the monomer and the kissing complex. The mean values (bars), including standard deviation (error bars) of at least three independent measurements, are depicted. The asterisks indicate the significance level: *P < 0.05, **P < 0.001 and ***P < 0.0001.
Characterization of the kissed loop region
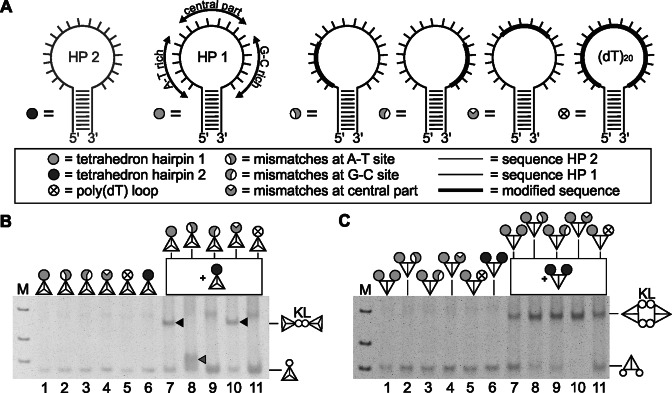
In canonical Watson–Crick duplexes mismatches reduce the double strand stability. The sequences of the loops of hairpin 1 and 2 are designed such that in principle kissing interaction could take place over the whole complementary loop region. However, it is not expected, that base pairing will include all nucleotides. We wanted to identify the favored hybridization sequence within the loop sequence. For that we designed three loop species of hairpin 1 for which we changed 6 nt at either one of the two lateral regions (the A-T rich and the G-C rich region) or the central loop region (Figure 9A). If those regions were involved in kissing interactions with hairpin 2 the changes would lead to mismatches, decreasing the stability of the kissing complex of the tetrahedrons with hairpin 2 and the tetrahedrons with hairpin 1 variants.
Figure 9.
Characterization of the kissing complex by mismatch analyses. (A) Schematic representation of the stem-loop sequences used for functionalization in this experiment. On the right next to hairpin 2: hairpin 1 with highlighted characteristics, on the right: hairpin with poly(dT) loop. Three further hairpins were designed, in which the loop sequence of hairpin 1 was modified in different regions. Six adjacent nucleotides at the A-T site, the G-C site or the central part of loop sequence hairpin 1 were substituted (bolt line) so that those sites can not participate in hydridization with hairpin 2. The schematic representation used in panels (B and C) is indicated. (B) The different monofunctionalized tetrahedrons were successfully assembled (lane 1–6). Each tetrahedron in lane 1–5 was incubated with tetrahedron hairpin 2 (lane 7–11). Resulting kissing-loop complexes are indicated by black triangles (lane 7 and 10). An intermediate possibly due to week kissing which migrates below the normal kissing complex band was visible after incubation of the tetrahedron with mismatches at the A-T rich site (gray triangle). (C) In an extended hybridizing experiment bifunctionalized tetrahedrons with different loop sequences as symbolized on top of the lanes were created and incubated with the bifunctionalized tetrahedron hairpin 2 (lane 6). While the potential interaction partners of the tetrahedron hairpin 2 had always one hairpin 1, the second was different as detailed in (A).
Indead, using mono-functionalized tetrahedrons, we could identify the lateral regions as important determinants for a stable kissing interaction, while changes in the sequence of the central part did even slightly enhance the stability of the kissing complex possibly by stabilizing the lateral hybridization (Figure 9B). This effect was also observed for an alternative mismatched central loop region (Supplementary Figure S7). If the A-T rich site was modified to be no longer involved in kissing annealing, the remaining G-C rich part led to the appearance of a structure, reminiscent of the intermediate seen in Figure 7A when the kissing complex was disassembled by temperature. No kissing annealing and no intermediate were observed for changes in the G-C-rich site, which might be due to the stronger interactions between guanine and cytosine compared to thymine and adenine.
Next we asked for the stability of kissing complexes formed by bifunctionalized tetrahedrons (at different vertices) with one original hairpin 1/hairpin 2 pair and one pair of hairpins as described before. In contrast to the first situation, the two possible kissing interactions might influence each other. We already showed, that while in general more kissing interactions increase the kissing complex stability, still, interactions involving different vertices (as compared to the same) empede effective kissing interaction of all stem-loops.
As expected from the results shown in Figure 9B, the highest yield of kissing complex was observable when a hairpin was used whose central part could not participate in base pairing (Figure 9C). This could be confirmed by using the alternative mismatched central loop region (Supplementary Figure S7) as described above. However, unexpectedly, the original kissing interaction (between hairpin 1 and 2) yielded less kissing product than the interaction involving one classical pair of hairpins and one pair with laterally changed sequences. Apparently, the interactions of the two fully complementary loop sequences negatively influenced each other, perhaps because less stable interactions were sustained. However, the downsizing of the complementary region at one loop in a way, that lateral kissing interaction was enforced, possibly favored a compact conformation that allowed the formation of a stabilized complex with all loops kissed simultaneously.
DISCUSSION
For the design of defined DNA nanostructures, it is essential to understand the properties of the used components. This opens the possibilities of controlling the assembly and final structure of the nanostructures and of applying such nanostructures for special purposes. We combined tetrahedrons with stem-loop structures to take advantage of the fact that the loop sequences can hybridize to each other if there are complementary sequences in the loop region. This non-canonical hybridization is called kissing-loop interaction. DNA tetrahedrons have been described and characterized in the past and show certain advantages for application in DNA nanotechnology. The synthesis of such tetrahedrons is relatively simple, e.g. the tetrahedrons can be assembled with four synthetic oligonucleotides, and the costs are low (8). As an advanced possibility, although not yet applied in this work, a tetrahedron could even be assembled from only one polynucleotide, which can be produced as single-stranded DNA in bacteria (35). Such biological synthesis might provide the opportunity for large-scale application of the modified tetrahedrons developed in this work. The tetrahedrons are robust DNA nanostructures. The lengths of the edges within the same tetrahedrons can be different (14), and even the length of an edge can be switched between two states (36). In our work, the robustness of the tetrahedral design could be confirmed by the observation that the modification of the constituting oligonucleotides, leading to the addition of one or more stem-loops, even at different vertices, still led to correct folding of the structure; this was demonstrated by the observations that the structure was ligatable and that the ligation product was not accessible to ExoIII digestion. However, so far, the usage of DNA tetrahedrons in contrast to, e.g. DNA origami (28), for nanotechnological purposes has been limited because only a few attempts have been made to assemble DNA tetrahedrons into larger structures. Such approaches also introduced free DNA ends into the tetrahedron sequence, possibly decreasing the stability of the tetrahedrons in the presence of environmental influences or exonucleolytic digestion.
Whereas hairpins are a common motif in RNA due to their single-stranded structure (e.g., in t-RNA), in DNA, the canonical Watson–Crick duplex is the dominant form. Natural DNA stem-loops are poorly understood but are known to play an important role in several cellular processes in prokaryotes (37), e.g. DNA hairpins in rolling-circle replication of the plasmid pT181 (37–39) as well as bacterial conjugation (40). DNA hairpins are also known to exist in bacteriophages, e.g. hairpin promoters regulating the transcription of the phage early genes (41,42), and as part of the primosome at the start of prokaryotic replication (37). Even in higher eukaryotes, the existence of DNA hairpins has been discussed. These hairpins might be the causative structure for triplet expansion diseases (43). DNA stem-loop structures either might be bound by regulatory proteins or might interact with each other (kissing stem-loops). Kissing-loop interaction has been characterized, e.g. in early Drosophila melanogaster embryo development, in which two mRNAs dimerize via two kissing-loop interactions (44). However, due to its importance for public health, the initial kissing-loop interaction of the RNA of the HI virus is the best-examined example of kissing-loop interaction. This initial loop–loop interaction results in final dimerization of the RNA genome of the virus. The structure of RNA kissing-loops and RNA–DNA kissing-loops have been analyzed by nuclear magnetic resonance (45–48), and the investigations have been extended by determining the structure of the homologous DNA–DNA kissing-loop (2). These investigations plus additional experimental approaches using model stem-loop structures and theoretical calculations (22–24,34) have led to the model of kissing-loop interaction as restricted to a short stretch of DNA. This phenomenon may be explained by the torsional stress of the annealing, which cannot be relaxed because in contrast to canonical annealing, no free DNA end is present. This implies that the kissing-loop complex is based on a weak interaction until the dimerization of the genome occurs, which is thermodynamically the much more favored form. This implication is also in line with the facts that hybridizing DNA loop regions longer than 14 base pairs promote hybridization efficiency and that two hairpins with long loop regions rapidly form long-lived kissing complexes (34). Furthermore, the topology of the DNA interaction in a kissing complex is completely different from the topology of linear DNA. For DNA hairpins, sequence-dependent formation of kissing complexes has been proposed. In the case of evenly distributed bases, kissing leads to the formation of a four-way junction (23), whereas a spatial separation of AT- and GC-rich regions allows lateral kissing interaction (24). We used the latter loop sequences in this work; kissing interaction in the GC-rich region within the loop might enable additional stabilizing base stacking (24,31). With respect to our experiments, this concept opens new possibilities and questions. If kissing-loop interaction is too weak, the complex of tetrahedrons could be unstable and dissociate too fast for nanotechnological applications. However, the reversibility of kissing-loop interaction might allow the disassembly and reassembly of tetrahedron kissing complexes under certain conditions, so it might be possible to design a tunable complex. Here, we demonstrated that number and position of kissing-loops, the temperature and the cation concentration as well as the loop sequence itself are useful parameters for this tuning, as discussed in the following paragraphs.
The first observation was that in our hands, kissing interaction of tetrahedrons resulted in a dimer as the main product, even for those tetrahedrons with higher numbers of stem-loops. As expected, our results show that the number of complementary loops that can participate in kissing interaction influences the yield and also the stability of the kissing-loop complex. The formation of a more distinct band for tetrahedrons with a higher number of loops hints at higher stability, such as during exposure to forces during electrophoresis. The higher yield of kissing-loop product when more loops are complementary to each other can be explained as follows. If, in case of two tetrahedrons with three complementary loops at one vertex, one pair of loops hybridizes, the hybridization of the other pairs might be facilitated, which would be a sort of cooperativity. Another explanation for the increased yield is that if one kissing-loop is melted, there might still be up to two loop–loop pairs holding the dimeric tetrahedron complex together, which increases the chance that the melted loop–loop pair is reconstituted. Therefore, the increased yield is the result of an increased efficiency of complex formation or higher stability of the assembled complex or both. For tetrahedrons with fewer stem-loops or with stem-loops located at separate vertices, the effect is reduced. Thus, it can be imagined that further combinations of numbers and positions of complementary or even non-complementary loops will further improve the tunable stability of kissing complexes.
Cations are regularly used in annealing reactions. In this context, we could demonstrate that Mg2+ is involved in the formation of a kissing-loop and also can be substituted with Ca2+ and Zn2+ (Figure 8). Mg2+, followed by Ca2+, promoted nucleic acid duplex stability. Purine bases were identified to be the preferential binding sites of Mg2+ (49) and were also part of the hybridizing loop sequence used in this study (22,24). In addition, the necessity of Mg2+ for kissing complexes has been well reported for RNA HIV-1 DIS stem-loop-based kissing complexes in the dimerization pathway of HIV RNA genomes (50,51) as well as for artificial RNA kissing complexes (52). It has also been reported that RNA kissing-loops display a stronger dependency on ionic strength than regular RNA helices do (53). The presence of Mg2+ was also demonstrated in the crystal structure of HIV RNA kissing complexes (32,54). With respect to potential nanotechnological applications, especially in the pharmaceutical area, the concentration of applied cations is important. Kissing-loops already exhibit reasonable stability at low ionic concentrations, such as in the blood. In human blood, the concentration of Mg2+ ranges from 0.65 to 1.05 mM (55), which is similar to the range that supports kissing-loop interaction in the case of the functionalized tetrahedrons presented here. For biomedical applications, a kissing complex might be used as a shuttle. For this purpose, the complex needs to be stable at body temperature (35–40°C) and needs to disassemble in a specific part of the body if treated with hyperthermia. We were able to establish a molecular species of tetrahedron kissing complexes (three complementary loops at three different vertices) in which, at normal body temperature, the complexes are fairly stable, whereas at an elevated temperature, decay of the complexes starts. Furthermore, it might be interesting to use kissing interaction instead of canonical annealing if heat-sensitive molecules, such as proteins, are to be caged within DNA nanostructures.
We also designed alternative loop sequences (Figure 9, Supplementary Figure S4) and could establish tetrahedrons with several loop pairs which could dimerize by different simultaneously kissed loops. Our results also show that the character of the loop sequence could effect the kissing efficiency. For weak kissing complexes formed by tetrahedrons with only one loop a completely complementary loop sequence was necessary to guarantee a stable kissing-loop dimer. Mismatches that knocked out interactions at lateral loop regions (the A-T rich or the G-C-rich sequence) led to a dramatically reduced stability of the kissing complex. Interestingly an altered sequence at the central loop regions not complementary to the interaction loop sequence did not effect kissing stability which is in line with recently published data for kissed DNA hairpins (23,24) and supports a more stable lateral hybridization. For firmer kissing complexes of tetrahedrons with more than one stem-loop at different vertices, which were eligible for caging of cargos, the effect of mismatches changed. We demonstrated this effect for tetrahedrons with two stem-loops where one loop was modified leading to mismatches. As discussed before kissing efficiency of tetrahedrons with stem-loops at different vertices was reduced in contrast to tetrahedrons with loops located nearby at one vertex. This effect was overcome by modifying the size of the complementary sequence within the loop region. This shows that the kissing modalities also depend on the manner how the nanostructures were functionalized by stem-loops. Therefore, fine tuning of the number, the position, as well as the character of the loop sequence and the resulting kissing complex might control the optimal stability and decay of drug-delivery tetrahedron complexes.
As mentioned before, kissing interaction of two loops does not include base pairing of the whole loop sequence, although total complementarity can exist. Non-annealed single-stranded regions within the loop enable the sequence-directed release of encapsulated cargos by strand invasion. This was demonstrated for one-looped tetrahedrons for which an invasion strand (capping oligonucleotide) hybridized to unpaired nucleotides of the loop sequence. The disassembly of the kissing complex was then effected by a slow strand displacement reaction. Although the kissing complex of one-loop tetrahedrons was the least stable tested complex, the disintegration of the kissing complex did not occur immediately. The above finding suggests that this weak kissing complex also had high stability. The slow disassembly by the capping oligonucleotide additionally enables fine tuning for the release of cargos adapted to the ambient medium (ionic strength and temperature). Thus, a tetrahedron kissing complex enclosing a cargo could first bind to special tissue, perhaps accelerated by the binding of proteins (17) and aptamers (56), and later on, the cargo could be liberated by disassembling the kissing complex by the addition of a capping oligonucleotide. The cargo could be transported inside the cavity between two tetrahedrons forming a kissing complex. The dimensions of the cavity might be adapted to the size of the encaged particle by changing the form of the tetrahedrons, e.g. the lengths of the edges (8) or the hairpin stems. Tetrahedrons modified with stem-loop sequences show further interesting properties which might be a valuable addition to the already existing nanotechnological toolbox. Due to the absence of 5′ and 3′ ends they should be resistant to exonucleolytic digestion (9) making them suitable for in vivo applications. The hybridization strength can be modified by the number and the sequence of stem-loops located at a certain vertex of a tetrahedron. Therefore the stability of the different kissing-loop complexes in a complex of two or more tetrahedrons can be variable and one can imagine that such a property can be developed into a nanotechnological mechanical element such as a hinge. In such constructs weaker hybridized kissing-loop complexes at one vertex might already melt under less stringent conditions whereas at another vertex (the hinge) the kissing complexes are more stable and therefore the kissing-loop complexes still exist. Such nanomechanical devices are interesting e.g. for nanobots. The described dependence of the hybridization of kissing-loop complexes on the presence of divalent cations and temperature might be used for the development of nanosensors especially if the tetrahedrons are labeled with fluorescent dyes allowing the read-out with the FRET technique. The described reversibility of kissing-loop interaction caused by the fact that in our system the hybridization of two complementary loop sequences is halted at the kissing-loop state and does not continue to the canonical double stranded longer DNA helix would allow to use the tetrahedrons as a part of a logical element (gate) without consumption of the used DNA structure allowing multiple rounds of computation. So several applications for the tetrahedron-stem-loop system can be imagined. The kissing-loop motif itself could also be a promising tool for the upgrade of nanobiotechnological molecules apart from the tetrahedron (e.g. cubes, octahedrons, DNA origami structures). In future experiments, several complementary loop–loop pairs with adapted loop sequences could be used for functionalization, opening the possibility of even more sophisticated complexes. Kissing-loop interaction provides moreover a simple, fast and easy way to assemble stable but modifiable structures in a one-pot reaction for carefully designed sequences.
Supplementary Material
Acknowledgments
We thank Holger Puchta for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation DFG [CFN project C5.4]. Funding for open access charge: Karlsruhe Institute of Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984;38:861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- 2.Barbault F., Huynh-Dinh T., Paoletti J., Lancelot G. A new peculiar DNA structure: NMR solution structure of a DNA kissing complex. J. Biomol. Struct. Dyn. 2002;19:649–658. doi: 10.1080/07391102.2002.10506771. [DOI] [PubMed] [Google Scholar]
- 3.Paillart J.C., Skripkin E., Ehresmann B., Ehresmann C., Marquet R. A loop-loop ‘kissing’ complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shetty S., Kim S., Shimakami T., Lemon S.M., Mihailescu M.R. Hepatitis C virus genomic RNA dimerization is mediated via a kissing complex intermediate. RNA. 2010;16:913–925. doi: 10.1261/rna.1960410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunel C., Marquet R., Romby P., Ehresmann C. RNA loop-loop interactions as dynamic functional motifs. Biochimie. 2002;84:925–944. doi: 10.1016/s0300-9084(02)01401-3. [DOI] [PubMed] [Google Scholar]
- 6.Cao S., Chen S.J. Predicting kissing interactions in microRNA-target complex and assessment of microRNA activity. Nucleic Acids Res. 2012;40:4681–4690. doi: 10.1093/nar/gks052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao F., Gulay S.P., Kasprzak W., Dinman J.D., Shapiro B.A., Simon A.E. The kissing-loop T-shaped structure translational enhancer of Pea enation mosaic virus can bind simultaneously to ribosomes and a 5′ proximal hairpin. J. Virol. 2013;87:11987–12002. doi: 10.1128/JVI.02005-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodman R.P., Berry R.M., Turberfield A.J. The single-step synthesis of a DNA tetrahedron. Chem. Commun. (Camb) 2004:1372–1373. doi: 10.1039/b402293a. [DOI] [PubMed] [Google Scholar]
- 9.Walsh A.S., Yin H., Erben C.M., Wood M.J., Turberfield A.J. DNA cage delivery to mammalian cells. ACS Nano. 2011;5:5427–5432. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 10.Lee H., Lytton-Jean A.K., Chen Y., Love K.T., Park A.I., Karagiannis E.D., Sehgal A., Querbes W., Zurenko C.S., Jayaraman M., et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford R., Erben C.M., Periz J., Hall L.M., Brown T., Turberfield A.J., Kapanidis A.N. Non-covalent single transcription factor encapsulation inside a DNA cage. Angew. Chem. Int. Ed. Engl. 2013;52:2284–2288. doi: 10.1002/anie.201207914. [DOI] [PubMed] [Google Scholar]
- 12.Kim K.R., Kim D.R., Lee T., Yhee J.Y., Kim B.S., Kwon I.C., Ahn D.R. Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells. Chem. Commun. (Camb) 2013;49:2010–2012. doi: 10.1039/c3cc38693g. [DOI] [PubMed] [Google Scholar]
- 13.Kim K.R., Lee Y.D., Lee T., Kim B.S., Kim S., Ahn D.R. Sentinel lymph node imaging by a fluorescently labeled DNA tetrahedron. Biomaterials. 2013;34:5226–5235. doi: 10.1016/j.biomaterials.2013.03.074. [DOI] [PubMed] [Google Scholar]
- 14.Goodman R.P., Schaap I.A., Tardin C.F., Erben C.M., Berry R.M., Schmidt C.F., Turberfield A.J. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 15.Mastroianni A.J., Claridge S.A., Alivisatos A.P. Pyramidal and chiral groupings of gold nanocrystals assembled using DNA scaffolds. J. Am. Chem. Soc. 2009;131:8455–8459. doi: 10.1021/ja808570g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erben C.M., Goodman R.P., Turberfield A.J. Single-molecule protein encapsulation in a rigid DNA cage. Angew. Chem. Int. Ed. Engl. 2006;45:7414–7417. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
- 17.Duckworth B.P., Chen Y., Wollack J.W., Sham Y., Mueller J.D., Taton T.A., Distefano M.D. A universal method for the preparation of covalent protein-DNA conjugates for use in creating protein nanostructures. Angew. Chem. Int. Ed. Engl. 2007;46:8819–8822. doi: 10.1002/anie.200701942. [DOI] [PubMed] [Google Scholar]
- 18.Simmons C.R., Schmitt D., Wei X., Han D., Volosin A.M., Ladd D.M., Seo D.K., Liu Y., Yan H. Size-selective incorporation of DNA nanocages into nanoporous antimony-doped tin oxide materials. ACS Nano. 2011;5:6060–6068. doi: 10.1021/nn2019286. [DOI] [PubMed] [Google Scholar]
- 19.Wilks T.R., Bath J., de Vries J.W., Raymond J.E., Herrmann A., Turberfield A.J., O'Reilly R.K. ‘Giant surfactants’ created by the fast and efficient functionalization of a DNA tetrahedron with a temperature-responsive polymer. ACS Nano. 2013;7:8561–8572. doi: 10.1021/nn402642a. [DOI] [PubMed] [Google Scholar]
- 20.Zhou T., Wang Y., Dong Y., Chen C., Liu D., Yang Z. Tetrahedron DNA dendrimers and their encapsulation of gold nanoparticles. Bioorg. Med. Chem. 2014;22:4391–4394. doi: 10.1016/j.bmc.2014.05.062. [DOI] [PubMed] [Google Scholar]
- 21.Xing S., Jiang D., Li F., Li J., Li Q., Huang Q., Guo L., Xia J., Shi J., Fan C., et al. Constructing higher-order DNA nanoarchitectures with highly purified DNA nanocages. ACS Appl. Mater. Interfaces. 2015;7:13174–13179. doi: 10.1021/am505592e. [DOI] [PubMed] [Google Scholar]
- 22.Bois J.S., Venkataraman S., Choi H.M., Spakowitz A.J., Wang Z.G., Pierce N.A. Topological constraints in nucleic acid hybridization kinetics. Nucleic Acids Res. 2005;33:4090–4095. doi: 10.1093/nar/gki721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romano F., Hudson A., Doye J.P., Ouldridge T.E., Louis A.A. The effect of topology on the structure and free energy landscape of DNA kissing complexes. J. Chem. Phys. 2012;136:215102-1–215102-9. doi: 10.1063/1.4722203. [DOI] [PubMed] [Google Scholar]
- 24.Sulc P., Romano F., Ouldridge T.E., Rovigatti L., Doye J.P., Louis A.A. Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 2012;137:135101. doi: 10.1063/1.4754132. [DOI] [PubMed] [Google Scholar]
- 25.Sanguinetti C.J., Dias Neto E., Simpson A.J. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques. 1994;17:914–921. [PubMed] [Google Scholar]
- 26.Lilley D.M. Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 27.Ke Y., Sharma J., Liu M., Jahn K., Liu Y., Yan H. Scaffolded DNA origami of a DNA tetrahedron molecular container. Nano Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 28.Rothemund P.W. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 29.Andersen E.S., Dong M., Nielsen M.M., Jahn K., Subramani R., Mamdouh W., Golas M.M., Sander B., Stark H., Oliveira C.L., et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 30.Sehdev P., Crews G., Soto A.M. Effect of helix stability on the formation of loop-loop complexes. Biochemistry. 2012;51:9612–9623. doi: 10.1021/bi300481v. [DOI] [PubMed] [Google Scholar]
- 31.Yakovchuk P., Protozanova E., Frank-Kamenetskii M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ennifar E., Walter P., Ehresmann B., Ehresmann C., Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 33.Cao S., Chen S.J. Structure and stability of RNA/RNA kissing complex: with application to HIV dimerization initiation signal. RNA. 2011;17:2130–2143. doi: 10.1261/rna.026658.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green S.J., Lubrich D., Turberfield A.J. DNA hairpins: fuel for autonomous DNA devices. Biophys. J. 2006;91:2966–2975. doi: 10.1529/biophysj.106.084681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Wei B., Nangreave J., Lin C., Liu Y., Mi Y., Yan H. A replicable tetrahedral nanostructure self-assembled from a single DNA strand. J. Am. Chem. Soc. 2009;131:13093–13098. doi: 10.1021/ja903768f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodman R.P., Heilemann M., Doose S., Erben C.M., Kapanidis A.N., Turberfield A.J. Reconfigurable, braced, three-dimensional DNA nanostructures. Nat. Nanotechnol. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 37.Bikard D., Loot C., Baharoglu Z., Mazel D. Folded DNA in action: hairpin formation and biological functions in prokaryotes. Microbiol. Mol. Biol. Rev. 2010;74:570–588. doi: 10.1128/MMBR.00026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.del Solar G., Giraldo R., Ruiz-Echevarria M.J., Espinosa M., Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan S.A. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid. 2005;53:126–136. doi: 10.1016/j.plasmid.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez-Perez B., Lucas M., Cooke L.A., Vyle J.S., de la Cruz F., Moncalian G. Analysis of DNA processing reactions in bacterial conjugation by using suicide oligonucleotides. EMBO J. 2007;26:3847–3857. doi: 10.1038/sj.emboj.7601806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai X., Rothman-Denes L.B. Sequence and DNA structural determinants of N4 virion RNA polymerase-promoter recognition. Genes Dev. 1998;12:2782–2790. doi: 10.1101/gad.12.17.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai X., Greizerstein M.B., Nadas-Chinni K., Rothman-Denes L.B. Supercoil-induced extrusion of a regulatory DNA hairpin. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2174–2179. doi: 10.1073/pnas.94.6.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volker J., Makube N., Plum G.E., Klump H.H., Breslauer K.J. Conformational energetics of stable and metastable states formed by DNA triplet repeat oligonucleotides: implications for triplet expansion diseases. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14700–14705. doi: 10.1073/pnas.222519799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrandon D., Koch I., Westhof E., Nusslein-Volhard C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dardel F., Marquet R., Ehresmann C., Ehresmann B., Blanquet S. Solution studies of the dimerization initiation site of HIV-1 genomic RNA. Nucleic Acids Res. 1998;26:3567–3571. doi: 10.1093/nar/26.15.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baba S., Takahashi K., Noguchi S., Takaku H., Koyanagi Y., Yamamoto N., Kawai G. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. 2005;138:583–592. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi K., Baba S., Hayashi Y., Koyanagi Y., Yamamoto N., Takaku H., Kawai G. NMR analysis of intra- and inter-molecular stems in the dimerization initiation site of the HIV-1 genome. J. Biochem. 2000;127:681–686. doi: 10.1093/oxfordjournals.jbchem.a022657. [DOI] [PubMed] [Google Scholar]
- 48.Collin D., van Heijenoort C., Boiziau C., Toulme J.J., Guittet E. NMR characterization of a kissing complex formed between the TAR RNA element of HIV-1 and a DNA aptamer. Nucleic Acids Res. 2000;28:3386–3391. doi: 10.1093/nar/28.17.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakano S., Fujimoto M., Hara H., Sugimoto N. Nucleic acid duplex stability: influence of base composition on cation effects. Nucleic Acids Res. 1999;27:2957–2965. doi: 10.1093/nar/27.14.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laughrea M., Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi K.I., Baba S., Chattopadhyay P., Koyanagi Y., Yamamoto N., Takaku H., Kawai G. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA. 2000;6:96–102. doi: 10.1017/s1355838200991635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horiya S., Li X., Kawai G., Saito R., Katoh A., Kobayashi K., Harada K. RNA LEGO: magnesium-dependent formation of specific RNA assemblies through kissing interactions. Chem. Biol. 2003;10:645–654. doi: 10.1016/s1074-5521(03)00146-7. [DOI] [PubMed] [Google Scholar]
- 53.Weixlbaumer A., Werner A., Flamm C., Westhof E., Schroeder R. Determination of thermodynamic parameters for HIV DIS type loop-loop kissing complexes. Nucleic Acids Res. 2004;32:5126–5133. doi: 10.1093/nar/gkh841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jossinet F., Paillart J.C., Westhof E., Hermann T., Skripkin E., Lodmell J.S., Ehresmann C., Ehresmann B., Marquet R. Dimerization of HIV-1 genomic RNA of subtypes A and B: RNA loop structure and magnesium binding. RNA. 1999;5:1222–1234. doi: 10.1017/s1355838299990982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saris N.E., Mervaala E., Karppanen H., Khawaja J.A., Lewenstam A. Magnesium. an update on physiological, clinical and analytical aspects. Clin. Chim. Acta. 2000;294:1–26. doi: 10.1016/s0009-8981(99)00258-2. [DOI] [PubMed] [Google Scholar]
- 56.Sun H., Zhu X., Lu P.Y., Rosato R.R., Tan W., Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol. Ther. Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.