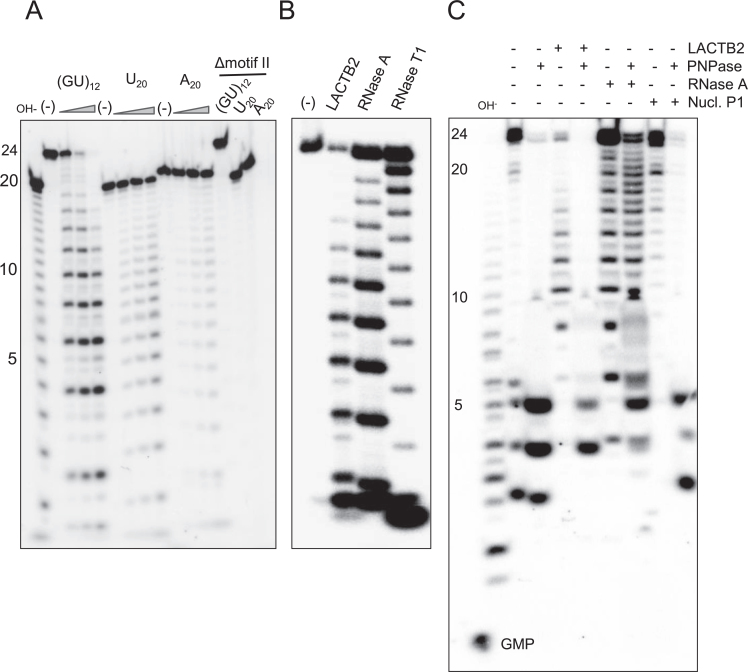
Figure 5.
Recombinant LACTB2 prefers U, but not G or poly(A). (A) LACTB2 was incubated for 15, 30 and 60 min, at 37°C with poly(GU)12, poly(U)20 or poly(A)20, in the presence of yeast tRNA. The RNA substrates were labeled with [32P] at the 5΄ end. Lane (L): Nucleotide ladder prepared by alkaline hydrolysis of poly(U)20. Lane (−): RNA substrate incubated with no protein for 60 min. Lanes under the triangle—incubation for 15, 30 and 60 min. Lanes labeled ΔmotifII: the RNAs were incubated for 60 min with the Δmotif II mutated protein in which the six amino acids of motif II were deleted (see Figure 7). Following incubation, the RNA was purified and analyzed by denaturing PAGE and autoradiography. (B) Poly(GU)12 was digested in the presence of yeast tRNA with LACTB2, RNase A (cleaves following U) and RNase T1 (cleaves following G). Lane (−) is the same as in panel A. (C) LACTB2 cleavage products have 3΄-OH termini and therefore are sensitive to digestion by the exoribonuclease PNPase. 5΄ [32P] (GU)12 RNA was digested with the enzymes as indicated on the top. When using two enzymes, the RNA was purified by phenol extraction and EtOH precipitation between the incubations. The full length 24 nt RNA contains a 3΄-OH and therefore is sensitive to PNPase. The cleavage products of RNase A have 3΄-phosphate while those of Nuclease P1 contain 3΄-OH. Some leakage of the digestion products of PNPase to the lane of no protein (−) happened when the gel was loaded with the samples. Nucl. P1: Nuclease P1.