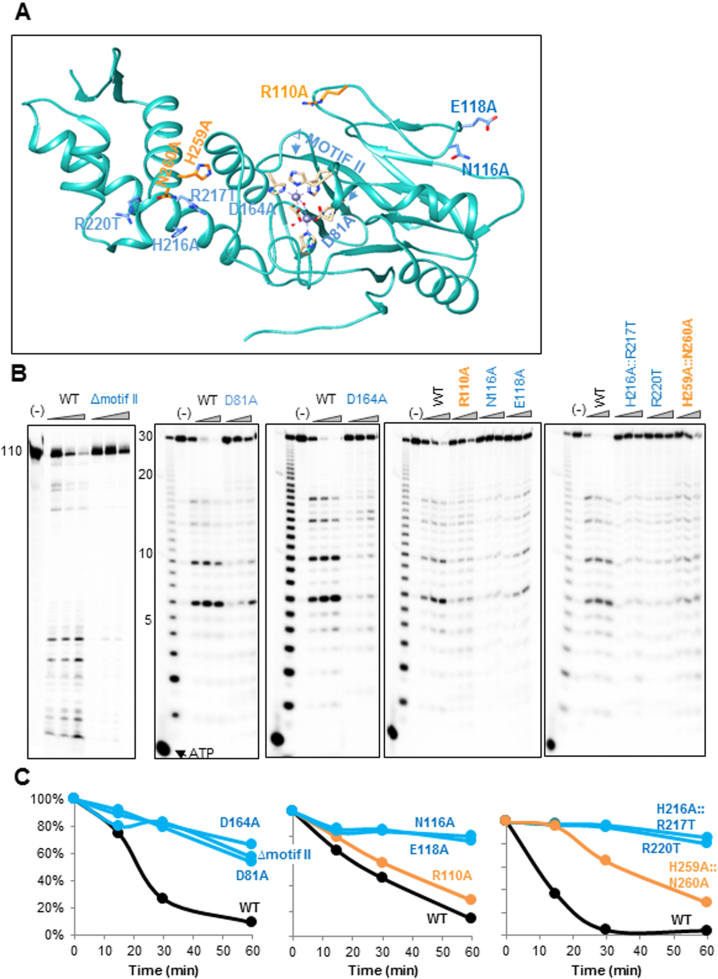
Figure 7.
Mutation of certain amino acids in the vicinity of the catalytic site, as well as at other locations, significantly inhibits the ribonucleolytic activity. (A) The locations of nine LACTB2 mutations introduced in this work are indicated in the structural image of the protein, created using Chimera. The mutated amino acids that significantly inhibited the endoribonucleolytic cleavage activity are colored in blue, and those not impairing the activity are colored in orange. The ΔMotif II mutation is a deletion of the entire motif II (amino acids HWHRDH). The other mutations, located in the vicinity of the cleavage catalytic site, were D81A and D164A. Additional mutated amino acids were: R110A, N116A, E118A, H216A::R217T, R220T and the double mutant H259A::N260A. The two zinc ions are presented as purple spheres and hydrogen bonds are presented as with purple dashed lines. (B) Non-mutated (WT) and LACTB2 mutants ΔMotif II, D81A, D164A, R110A, N116A, E118A, H216A::R217T, R220T, H259A::N260A were incubated with 5΄ [32P] −110 nt RNA corresponding to the mitochondrial COX1 transcript (left panel) or 30 nt RNA described in Figure 4 at 37°C, followed by the addition of formamide dye and analysis by denaturing gel and autoradiography. Deca RNA Marker was fractionated in the first lane to the right and the nucleotide ladder of the 30 nt RNA substrate is shown in the next lane. Lane (−): RNA incubation for 60 min with no added protein. Lanes under the triangle: incubation for 15, 30 and 60 min. (C) Kinetic graphs displaying the quantification of the disappearance of the full-length substrate RNA, when incubated with LACTB2 WT or the mutated proteins. The amount of RNA present in the lane marked (−) was taken as 100%. WT is colored in black, RNA cleavage in mutants displaying similar activity to WT is colored in orange. The activity of mutated proteins in which a significant inhibition was observed is colored in blue.