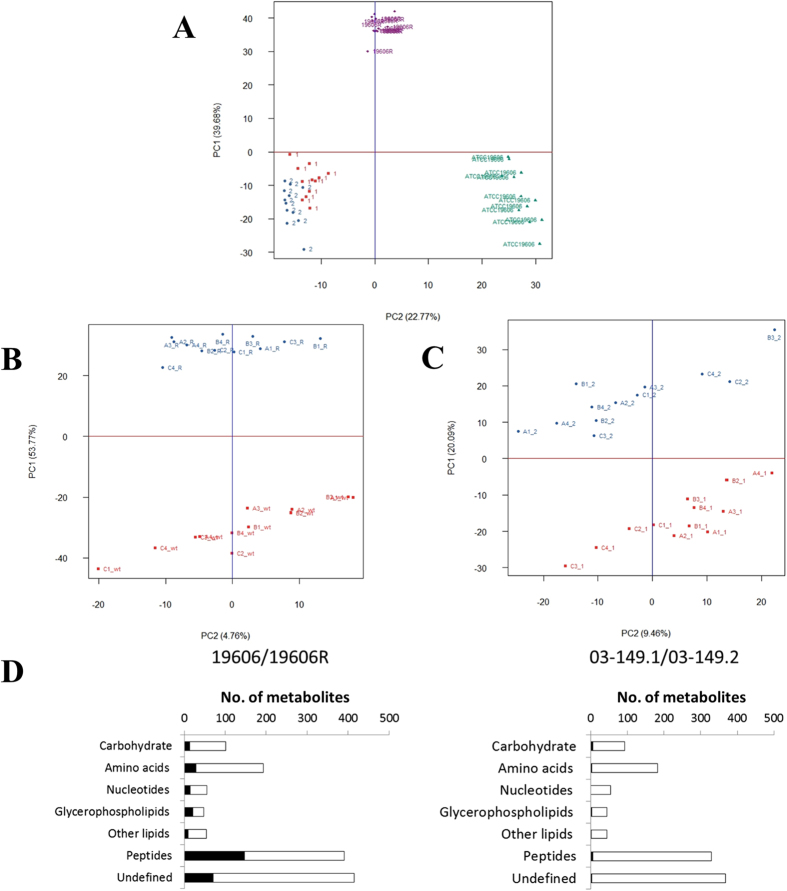
Figure 3.
(A) PCA score plot of four A. baumannii strains. (B) PCA score plot of paired polymyxin-resistant 19606R and the wild-type ATCC 19606. (C) PCA score plot of paired polymyxin-resistant 03–149.2 and polymyxin-susceptible 03–149.1 clinical isolates. Each data set for individual strains represents a total of 12 sample replicates (3 biological replicates and each with 4 technical replicates). (D) Pathway-focused representation of the significant metabolites (black bars) and total number of putatively identified metabolites (open bars) for the polymyxin-resistant 19606R relative to the wild-type ATCC 19606 (left) and the polymyxin-resistant clinical isolate 03–149.2 relative to the polymyxin-susceptible isolate 03–149.1 (right). Significant metabolites were selected by at least 2-fold difference (p < 0.05) in the metabolites levels of polymyxin-resistant strain relative to the polymyxin-susceptible strain.