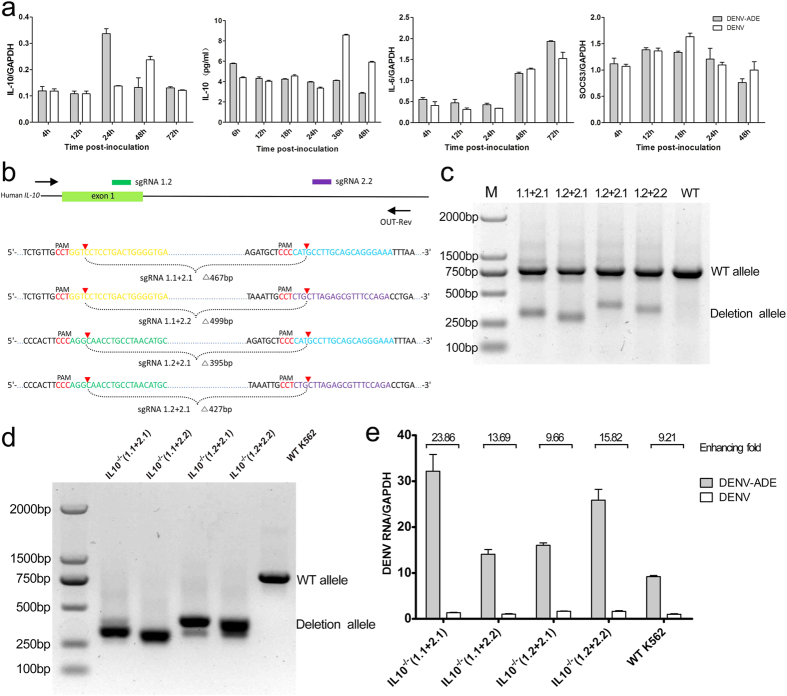
Figure 5.
DENV-ADE infection of K562 is independent of IL-10 function (a) K562 cells were infected with DENV alone or DENV-antibody complex or were mock infected. Cellular RNA was extracted and use for gene expression quantification of IL-10, IL-6 and SOCS3 using qRT-PCR; the supernatants of cell cultures were tested for IL-10 using an ELISA kit. N = 3; error bars show the means ± SEM. (b) Four pairs of sgRNAs were designed to target the human IL-10 loci. Target sequences and PAMs (red) are shown in respective colours, and sites of cleavage by Cas9 are indicated by red triangles. A predicted junction is shown below. (c) Cell populations transfected with paired sgRNAs (1.1 + 2.1, 1.1 + 2.2, 1.2 + 2.1, 1.2 + 2.2) were assayed by PCR (Out-Fwd and Out-Rev primers were used), reflecting a deletion of ~270 bp. (d) Paired sgRNA transfected cells were clonally isolated and expanded for genotyping analysis of deletions using PCR (using the Out-Fwd and Out-Rev primers) and sequencing. Each isolate of K562 with homozygous deletions was subjected to DENV-ADE/DENV infections. (e) Cellular dengue genome RNA was quantified using qRT-PCR with GAPDH as an internal control; the fold enhancement in each K562 isolate was expressed as a ratio of DENV/DENV-ADE. N = 3; error bars show the means ± SEM.