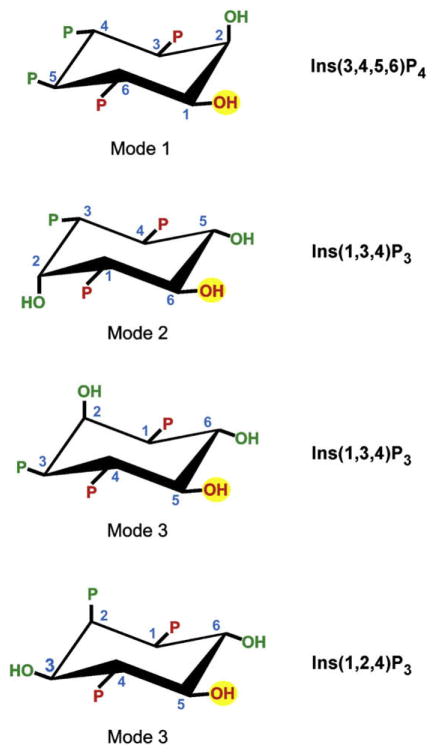
Fig. 4.
A model for the structural determinants of ligand specificity for mamalian ITPK1. The figure depicts our proposal (Ho et al., 2002; Riley et al., 2006) that there are three modes of binding of inositol phosphates to mamalian ITPK1. It can be illuminating to consider these different binding modes (i.e., “1”, “2” and “3”) as permitting 1-kinase, 6-kinase and 5-kinase activities, respectively. These phosphorylation sites are marked with a yellow circle. Three groups in Ins(3,4,5,6)P4, Ins(1,3,4)P3 and Ins(1,2,4)P3 (coloured red) are conserved in all three of these proposed binding modes. We have previously noted that these groups by themselves are insufficient to designate substrate specificity, so we have proposed a combinatorial recognition model in which some of the additional groups (coloured green) contribute to ligand recognition, but in a mode-specific manner.