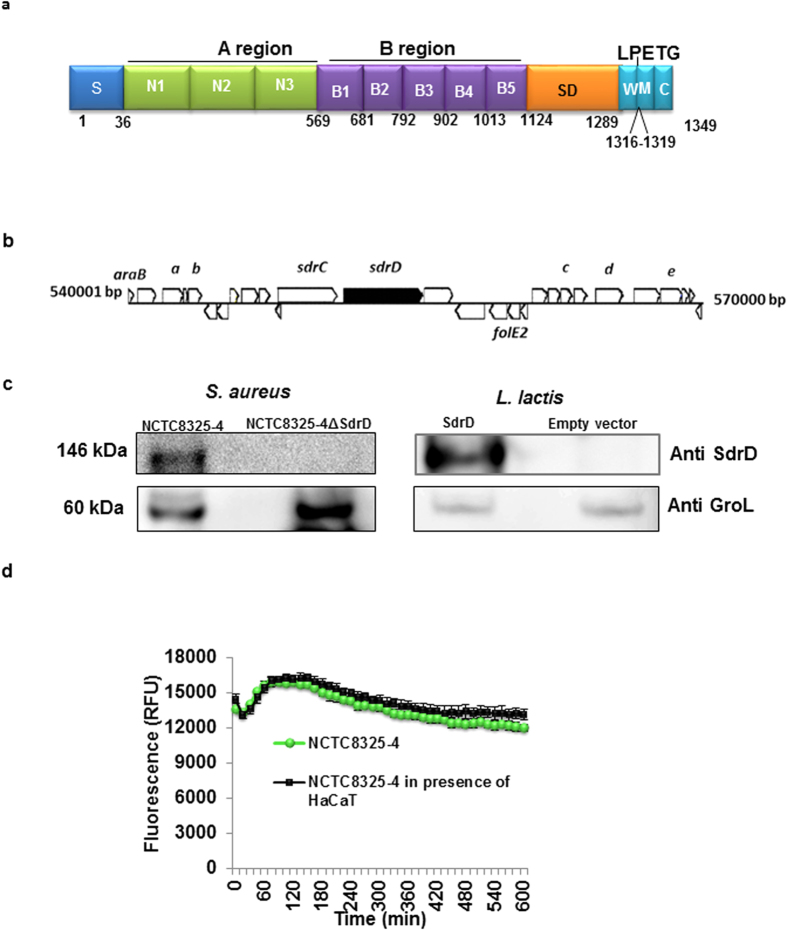
Figure 1. sdrD gene localization and expression in S. aureus NCTC8325-4.
(a) Schematic representation of SdrD domain structure in S. aureus NCTC8325-4 based on UniProtKB. S, signal sequence; A region composed of N1, N2 and N3; B repeats composed of B1 to B5; SD, serine-aspartate acid repeat region; W, wall-spanning fragment; LPETG, cell wall anchoring motif; M, transmembrane domain; C, cytoplasmic domain. (b) sdrC and sdrD is located between ORFs encoding hypothetical proteins according to annotation is from KEGG Genome map. Gene and protein name based on UniProtKB: araB, ribonuclokinase; a: SAOUHSC_00536, Branched-chain-amino-acid aminotransferase; b: SAOUHSC_00538, Haloacid dehalogenase-like hydrolase; sdrC, serine-aspartate repeat-containing protein C; sdrD, serine-aspartate repeat-containing protein D; folE2, GTP cyclohydrolase FolE2; c: SAOUHSC_00554, SIS domain protein; d: SAOUHSC_00556, Proline/betaine transporter; e: SAOUHSC_00558, Acetyl-CoA acetyltransferase (c) Immunoblot using SdrD A-region and GroL antibodies on cell lysate S. aureus NCTC8325-4 or its isogenic mutant NCTC8325-4ΔsdrD and L.lactis with pMG36e-SdrD (SdrD) or pMG36e (empty vector). (d) sdrD promoter activity in DMEM supplemented with FBS without agitation in the absence ( ) or presence (■) of HaCaT cells using S. aureus NCTC8325-4 harbouring sdrD-GFP reporter construct. Data expressed as mean ± standard deviation (SD) of an individual experiment.
) or presence (■) of HaCaT cells using S. aureus NCTC8325-4 harbouring sdrD-GFP reporter construct. Data expressed as mean ± standard deviation (SD) of an individual experiment.